













| General Information: |
|
| Name(s) found: |
GACP3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 838 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear envelope
[IDA]
internal side of plasma membrane [IDA] |
| Biological Process: |
microtubule nucleation
[TAS]
|
| Molecular Function: |
tubulin binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEDDDQQKAA DLVQELVLRL VSQNPQTPNL DPNSPAFLKT LRYAFRILSS RLTPSVLPDA 60
61 TAIAESLKRR LATQGKSSDA LAFADLYTKF ASKTGPGSVN NKWALVYLLK IVSDDRKSAI 120
121 NGLDSSVLLP NLGIGDTGNG VLSRGEAKKK DWSNGVLLVS KDPENLRDIA FREYAILVKE 180
181 ENEVTEEVLV RDVLYASQGI DGKYVKFNSE IDGYAVQESV KVPRATRIMV RMLSELGWLF 240
241 RKVKTFITES MDRFPAEDVG TVGQAFCAAL QDELSDYYKL LAVLEAQAMN PIPLVSESAS 300
301 SNNYLSLRRL SVWFAEPMVK MRLMAVLVDK CKVLRGGAMA GAIHLHAQHG DPLVHDFMMS 360
361 LLRCVCSPLF EMVRSWVLEG ELEDTFGEFF VVGQPVKVDL LWREGYKLHP AMLPSFISPS 420
421 LAQRILRTGK SINFLRVCCD DHGWADAASE AAAASGTTTR RGGLGYGETD ALEHLVTEAA 480
481 KRIDKHLLDV LYKRYKFKEH CLAIKRYLLL GQGDFVQYLM DIVGPKLSEP ANNISSFELA 540
541 GFLEAAIRAS NAQYDDRDML DRLRVKMMPH GSGDRGWDVF SLEYEARVPL DTVFTESVLS 600
601 KYLRVFNFLW KLKRVEHALI GIWKTMKPNC ITSNSFVKLQ SSVKLQLLSA LRRCQVLWNE 660
661 MNHFVTNFQY YIMFEVLEVS WSNFSKEMEA AKDLDDLLAA HEKYLNAIVG KSLLGEQSQT 720
721 IRESLFVLFE LILRFRSHAD RLYEGIHELQ IRSKESGREK NKSQEPGSWI SEGRKGLTQR 780
781 AGEFLQSMSQ DMDSIAKEYT SSLDGFLSLL PLQQSVDLKF LFFRLDFTEF YSRLHSKG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
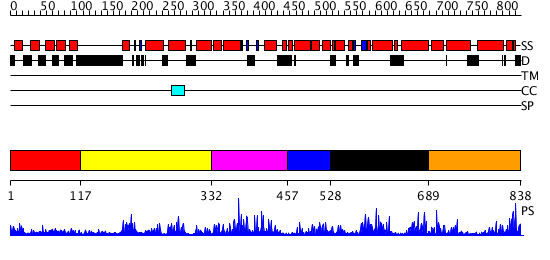
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | 1.094983 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [117..331] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [332..456] | 2.199989 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [457..527] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [528..688] | 1.204993 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [689..838] | 2.192989 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)