













| General Information: |
|
| Name(s) found: |
gi|7485475
[NCBI NR]
gi|3337353 [NCBI NR] gi|15226250 [NCBI NR] |
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 718 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
embryonic development ending in seed dormancy
[IMP]
pollen development [IMP] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMMKSTDESE TKWKIDDAAE LARDFWLERD GISALKLTEK TISDHGNHEK CSSHHELQGD 60
61 IFYAQAKKTD DTDLKCLYLF ASVDAYSMAT LLSPNALTLR SFRGYARSLI KLGDQLRINK 120
121 FYEKAVSKAK LGMSITSPQG KSTDGGYFTK IKTELQNWID LATTKMQATV AVSDIGTMDN 180
181 QKEPGEEDTS FDRLKKIWVK LDDKTKINFL VVDFEKLIGY IQDKHGTEVK EHFQKCIPIA 240
241 NNLRWRCWKC HICSQVNYCF TDCKMHILDN HVHNSEPDFS ARPKCVDEIL ADMICCGDWK 300
301 PVDTAEAENL IKDRAKRGEE FVYVNGWCSD WPEAKDRERE NVLKQFAEVL KSSCAKENCT 360
361 LSCTLWDWLR DYTEENLELP GVPGIYLDQC CFFKNPQCIC FLDLKHLEHI LKYFRQLTTD 420
421 VRASLVPKVV NQFWENSQVK ERIDLEGVAT YNLLLDKRLL YEEELESDKI GTVEHYKSTG 480
481 IFEDVMPKGD KIVSWILDCP EIDREFVSQM ANGVHNSKLW LAALRIVRCV IRKKESYYDK 540
541 RRKMLTYEKM LGEAETICDR EDTRKNVNQR STYESALRMK CEDLVGKQDD DTKCFLSVVR 600
601 DVFERQSSPS FEVFEDKKCI SELSSTVPND DVKKSLLTLR KSLKEKFPLI DSKILQNKST 660
661 YEKLIDVFPK LSAVEYRLVV LPFVKKFLQD KLKKMMKTNS SSVAAGDSAE RKPKKRHL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
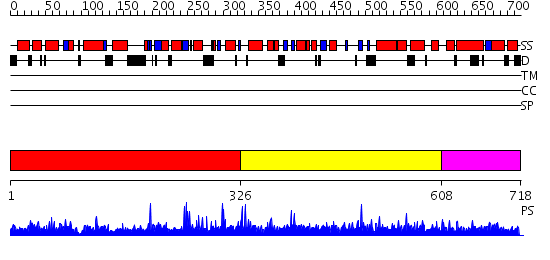
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..325] | 1.31 | Crystal structure of the CHIP U-box E3 ubiquitin ligase | |
| 2 | View Details | [326..607] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [608..718] | 1.016981 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)