













| General Information: |
|
| Name(s) found: |
FAB1B_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1791 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA]
|
| Biological Process: |
signal transduction
[ISS]
|
| Molecular Function: |
zinc ion binding
[ISS]
1-phosphatidylinositol-4-phosphate 5-kinase activity [ISS] phosphoinositide binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGTRDSNNRT FSEIVGLIKS WLPWRSEPAT VSRDFWMPDQ SCRVCYECDC QFTLINRRHH 60
61 CRHCGRVFCG KCTANSIPFA PSDLRTPRED WERIRVCNYC FRQWEQGDGG PHVSNITELS 120
121 TSPSETSLLS SKTSTTANSS SFALGSMPGL IGLNQRVHHG SDVSLHGVSS METSVTKQGK 180
181 ETSRRSSFIA TDVEDPSRFA LNSIRSDDEY DEYGAYQTDI ETSHSPRANE YYGPMEYNGM 240
241 GIDDVPCKHL GGETADQKSL SGSPLIHQCL ESLIREGSEQ FQKKSEHDGR DECEASSPAD 300
301 ISDDQVVEPV DFENNGLLWV PPEPENEEDE RESALFDEED NEGDASGEWG YLRPSTSFGS 360
361 GEYRGEDRTT EEHKKAMKNV VDGHFRALLA QLLQVENISV SDEEGKESWL EIITSLSWEA 420
421 ANLLKPDMSK SGGMDPGGYV KVKCLASGFR HDSMVVKGVV CKKNVVNRRM STKIEKARLL 480
481 ILGGGLEYQR VSNQLSSFDT LLQQEKDHLK MAVAKIHAER PNILLVEKSV SRFAQEYLLA 540
541 KDISLVLNIK RPLLDRIARC TGAQIIPSVD HLSSQKLGYC ENFRVDRYPE EHGSTGQVGK 600
601 KVVKTLMYFE HCPKPLGFTI LLRGANEDEL KKVKHVVQYG VFAAYHLALE TSFLADEGAS 660
661 PELPLNSPIT VALPDKSTSI ERSISTVPGF TVSTYEKSPT MLSCAEPQRA NSVPVSELLS 720
721 TTTNLSIQKD IPPIPYGSGW QAREINPSFV FSRHNISLNL PDRVIESRNS DLSGRSVPVD 780
781 TPADKSNPIV VADETTNNSL HLSGQGFVRK SSQIGTSIMV ENQDNGSELT IAQQQNNEKP 840
841 KETQSQKEEF PPSPSDHQSI LVSLSSRSVW KGTVCERSHL FRIKYYGSFD KPLGRFLRDH 900
901 LFDQSYRCRS CEMPSEAHVH CYTHRQGSLT ISVKKLQDYL LPGEKEGKIW MWHRCLRCPR 960
961 LNGFPPATLR VVMSDAAWGL SFGKFLELSF SNHAAASRVA CCGHSLHRDC LRFYGFGNMV1020
1021 ACFRYATIDV HSVYLPPSIL SFNYENQDWI QRETDEVIER AELLFSEVLN AISQIAEKGF1080
1081 RRRIGELEEV LQKEKAEFEE NMQKILHREV NEGQPLVDIL ELYRIHRQLL FQSYMWDHRL1140
1141 INASTLHKLE NSDDTKREEN EKPPLAKSQT LPEMNAGTNS LLTGSEVNLN PDGDSTGDTG1200
1201 SLNNVQKEAD TNSDLYQEKD DGGEVSPSKT LPDTSYPLEN KVDVRRTQSD GQIVMKNLSA1260
1261 TLDAAWIGER QTSVEIPTNN KVSLPPSTMS NSSTFPPISE GLMPIDLPEQ QNEFKVAYPV1320
1321 SPALPSKNYE NSEDSVSWLS VPFLNFYRSI NKNFLLSSQK LDTFGEHSPI YISSFREAEL1380
1381 QGGPRLLLPV GLNDIVVPVY DDEPTSMIAY ALMSPEYQRQ TSAEGESLVS YPSELNIPRP1440
1441 VDDTIFDPSR SNGSVDESIL SISSSRSTSL LDPLSYTKAL HARVSYGEDG TLGKVKYTVT1500
1501 CYYAKRFEAL RGICLPSELE YIRSLSRCKK WGAQGGKSNV FFAKTLDDRF IIKQVTKTEL1560
1561 ESFIKFAPAY FKYLSESIST KSPTCLAKIL GIYQVATKQL KSGKETKMDV LIMENLLFGR1620
1621 TVKRLYDLKG SSRARYNPDS SGSNKVLLDQ NLIEAMPTSP IFVGNKAKRL LERAVWNDTA1680
1681 FLALGDVMDY SLLVGVDEEK NELVLGIIDF LRQYTWDKHL ESWVKFTGIL GGPKNEAPTV1740
1741 ISPKQYKRRF RKAMTTYFLM VPDQWSPPNV VANNSKSDQP EETSQAGTQA E |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
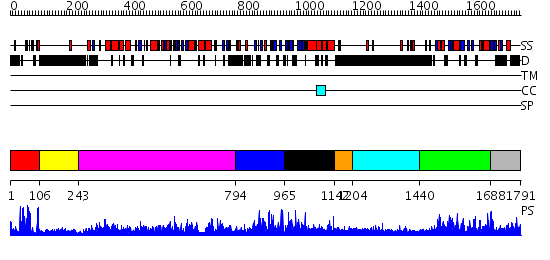
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..105] | 22.39794 | Eea1; Eea1 homodimerisation domain | |
| 2 | View Details | [106..242] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [243..793] | 151.0 | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) | |
| 4 | View Details | [794..964] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [965..1141] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1142..1203] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1204..1439] | 3.223996 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1440..1687] | 76.39794 | Phosphatidylinositol phosphate kinase IIbeta, PIPK IIbeta | |
| 9 | View Details | [1688..1791] | 3.14 | No description for 2gk9A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. | ||||||
| 6 | No functions predicted. | ||||||
| 7 | No functions predicted. | ||||||
| 8 |
|
||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)