













| General Information: |
|
| Name(s) found: |
gi|7268250
[NCBI NR]
gi|5302780 [NCBI NR] gi|25289826 [NCBI NR] gi|18414365 [NCBI NR] |
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 687 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
negative regulation of catalytic activity
[IEA]
proteolysis [IEA][ISS] |
| Molecular Function: |
identical protein binding
[IEA]
serine-type endopeptidase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGALPSKISY SPMSHHQNIL QEVIESSSVE DYLVRSYGRS FNGFAAKLTE SEKDKLIGME 60
61 GVVSVFPSTV YKLFTTRSYE FMGLGDKSNN VPEVESNVIV GVIDGGIWPE SKSFSDEGIG 120
121 PIPKKWKGTC AGGTNFTCNR KVIGARHYVH DSARDSDAHG SHTASTAAGN KVKGVSVNGV 180
181 AEGTARGGVP LGRIAVYKVC EPLGCNGERI LAAFDDAIAD GVDVLTISLG GGVTKVDIDP 240
241 IAIGSFHAMT KGIVTTVAVG NAGTALAKAD NLAPWLISVA AGSTDRKFVT NVVNGDDKML 300
301 PGRSINDFDL EGKKYPLAYG KTASNNCTEE LARGCASGCL NTVEGKIVVC DVPNNVMEQK 360
361 AAGAVGTILH VTDVDTPGLG PIAVATLDDT NYEELRSYVL SSPNPQGTIL KTNTVKDNGA 420
421 PVVPAFSSRG PNTLFSDILS NEHSKRNNRP MSQYISSIFT TGSNRVPGQS VDYYFMTGTS 480
481 MACPHVAGVA AYVKTLRPDW SASAIKSAIM TTAWAMNASK NAEAEFAYGS GFVNPTVAVD 540
541 PGLVYEIAKE DYLNMLCSLD YSSQGISTIA GGTFTCSEQS KLTMRNLNYP SMSAKVSASS 600
601 SSDITFSRTV TNVGEKGSTY KAKLSGNPKL SIKVEPATLS FKAPGEKKSF TVTVSGKSLA 660
661 GISNIVSASL IWSDGSHNVR SPIVVYT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
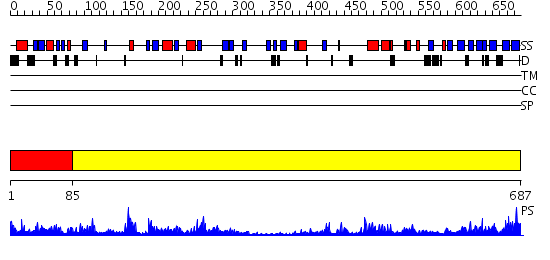
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | 37.154902 | Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin | |
| 2 | View Details | [85..687] | 47.154902 | Structure of C5a peptidase- a key virulence factor from Streptococcus |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)