













| General Information: |
|
| Name(s) found: |
VAR3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 758 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] |
| Biological Process: |
chloroplast organization
[IMP]
|
| Molecular Function: |
zinc ion binding
[IEA]
binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNNSTRLISL FSPHPPPLFL LRGLYISRIA NLRRFHRRAF PPSSVASTNL CSFRPLVSLP 60
61 PLIPTFPIGR FYNHQVRVSA ADFVPSYHNQ QLPEWTELLQ SLSKAGYFSD SGSISGLESE 120
121 FFPGFPDELL RPALACLALA RERPELLEML SRRDVEVLVE NGKPFLFKTG PDSLKRMSLY 180
181 LRSGLQGIGK LMDMEKASTV DLMRLILSYV VDVASSEESK QHNKEIMESS VRSLLSQIAK 240
241 MSLRPPESNV HDTMQNQYSD RDGQGVRSFQ NNVEMKRGDW ICSRCSGMNF ARNVKCFQCD 300
301 EARPKRQLTG SEWECPQCDF YNYGRNVACL RCDCKRPRDS SLNSANSDYS SDPELERRLV 360
361 ENEKKAQRWL SKVAQGGSDA NSVDTDEDFP EIMPLRKGVN RYVVSTRKPP LERRLANTEN 420
421 RVATDGNSKR SDDNALGSKT TRSLNEILGS SSSLTSRSDD KNVSSRRFES SQGINTDFVP 480
481 FVPLPSDMFA KKPKEETQIG LIDNIQVDGF SGGNQNVYQE DKSDANHSGK ETDRLEKEDH 540
541 KSEEPARWFK RVTELHNVSD LESAIPQEIS PEKMPMRKGE NRFVVSRKKD RSLTSPAYKR 600
601 PEDSDFVPFV PFPPDYFAKE KQPKESIDTL PAPATENVSQ VVQQEPREPS INKSDTVAVK 660
661 IRNGKSLEGS LVKESDLLDM SEEAKAERWF KRVAEIKNIS ELSEIPDEDF PSIMPMRKGV 720
721 NRFVVSKRKT PLERRLTSQR HQRNPHITNS DPTGKGDK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
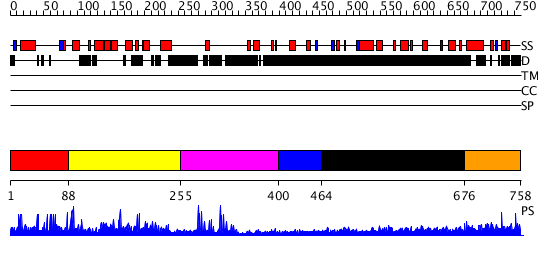
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [88..254] | 2.090968 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [255..399] | 14.178994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [400..463] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [464..675] | 4.173996 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [676..758] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.72 |
Source: Reynolds et al. (2008)