













| General Information: |
|
| Name(s) found: |
HFA4C_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 345 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
root development
[IMP]
response to heat [IEP] positive gravitropism [IMP] fruit morphogenesis [IMP] response to auxin stimulus [IMP] lateral root development [IMP] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDENNGGSSS LPPFLTKTYE MVDDSSSDSV VAWSENNKSF IVKNPAEFSR DLLPRFFKHK 60
61 NFSSFIRQLN TYGFRKVDPE KWEFLNDDFV RGRPYLMKNI HRRKPVHSHS LVNLQAQNPL 120
121 TESERRSMED QIERLKNEKE GLLAELQNQE QERKEFELQV TTLKDRLQHM EQHQKSIVAY 180
181 VSQVLGKPGL SLNLENHERR KRRFQENSLP PSSSHIEQVE KLESSLTFWE NLVSESCEKS 240
241 GLQSSSMDHD AAESSLSIGD TRPKSSKIDM NSEPPVTVTA PAPKTGVNDD FWEQCLTENP 300
301 GSTEQQEVQS ERRDVGNDNN GNKIGNQRTY WWNSGNVNNI TEKAS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
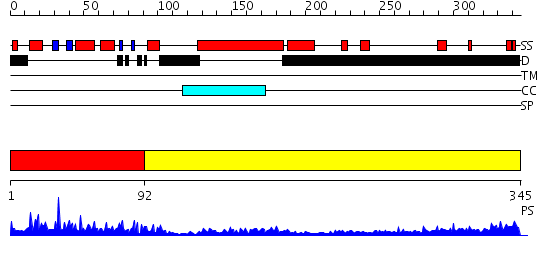
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..91] | 33.0 | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | |
| 2 | View Details | [92..345] | 1.46 | Ribonuclease domain of colicin E3; Colicin E3 translocation domain; Colicin E3 receptor domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)