













| General Information: |
|
| Name(s) found: |
RGA_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 587 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][TAS]
|
| Biological Process: |
negative regulation of gibberellic acid mediated signaling pathway
[IGI]
hyperosmotic salinity response [IGI] negative regulation of seed germination [IGI] response to abscisic acid stimulus [IGI] response to salt stress [IGI] response to far red light [IEP] regulation of oxygen and reactive oxygen species metabolic process [IGI] regulation of protein catabolic process [IMP] gibberellic acid mediated signaling pathway [TAS] regulation of transcription [TAS] jasmonic acid mediated signaling pathway [IGI] response to ethylene stimulus [IGI] |
| Molecular Function: |
transcription factor activity
[IDA][ISS][TAS]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRDHHQFQG RLSNHGTSSS SSSISKDKMM MVKKEEDGGG NMDDELLAVL GYKVRSSEMA 60
61 EVALKLEQLE TMMSNVQEDG LSHLATDTVH YNPSELYSWL DNMLSELNPP PLPASSNGLD 120
121 PVLPSPEICG FPASDYDLKV IPGNAIYQFP AIDSSSSSNN QNKRLKSCSS PDSMVTSTST 180
181 GTQIGGVIGT TVTTTTTTTT AAGESTRSVI LVDSQENGVR LVHALMACAE AIQQNNLTLA 240
241 EALVKQIGCL AVSQAGAMRK VATYFAEALA RRIYRLSPPQ NQIDHCLSDT LQMHFYETCP 300
301 YLKFAHFTAN QAILEAFEGK KRVHVIDFSM NQGLQWPALM QALALREGGP PTFRLTGIGP 360
361 PAPDNSDHLH EVGCKLAQLA EAIHVEFEYR GFVANSLADL DASMLELRPS DTEAVAVNSV 420
421 FELHKLLGRP GGIEKVLGVV KQIKPVIFTV VEQESNHNGP VFLDRFTESL HYYSTLFDSL 480
481 EGVPNSQDKV MSEVYLGKQI CNLVACEGPD RVERHETLSQ WGNRFGSSGL APAHLGSNAF 540
541 KQASMLLSVF NSGQGYRVEE SNGCLMLGWH TRPLITTSAW KLSTAAY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
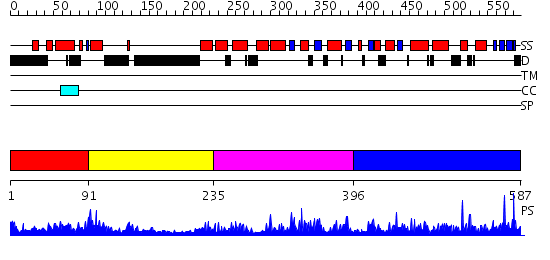
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..90] | 1.022992 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [91..234] | 2.119993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [235..395] | 2.129977 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [396..587] | 5.135963 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)