













| General Information: |
|
| Name(s) found: |
THIC_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 644 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] plastid [IDA] |
| Biological Process: |
detection of bacterium
[IMP]
response to vitamin B1 [IEP] thiamin biosynthetic process [IMP][ISS] |
| Molecular Function: |
ADP-ribose pyrophosphohydrolase activity
[IGI]
catalytic activity [ISS] iron-sulfur cluster binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAASVHCTLM SVVCNNKNHS ARPKLPNSSL LPGFDVVVQA AATRFKKETT TTRATLTFDP 60
61 PTTNSERAKQ RKHTIDPSSP DFQPIPSFEE CFPKSTKEHK EVVHEESGHV LKVPFRRVHL 120
121 SGGEPAFDNY DTSGPQNVNA HIGLAKLRKE WIDRREKLGT PRYTQMYYAK QGIITEEMLY 180
181 CATREKLDPE FVRSEVARGR AIIPSNKKHL ELEPMIVGRK FLVKVNANIG NSAVASSIEE 240
241 EVYKVQWATM WGADTIMDLS TGRHIHETRE WILRNSAVPV GTVPIYQALE KVDGIAENLN 300
301 WEVFRETLIE QAEQGVDYFT IHAGVLLRYI PLTAKRLTGI VSRGGSIHAK WCLAYHKENF 360
361 AYEHWDDILD ICNQYDVALS IGDGLRPGSI YDANDTAQFA ELLTQGELTR RAWEKDVQVM 420
421 NEGPGHVPMH KIPENMQKQL EWCNEAPFYT LGPLTTDIAP GYDHITSAIG AANIGALGTA 480
481 LLCYVTPKEH LGLPNRDDVK AGVIAYKIAA HAADLAKQHP HAQAWDDALS KARFEFRWMD 540
541 QFALSLDPMT AMSFHDETLP ADGAKVAHFC SMCGPKFCSM KITEDIRKYA EENGYGSAEE 600
601 AIRQGMDAMS EEFNIAKKTI SGEQHGEVGG EIYLPESYVK AAQK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
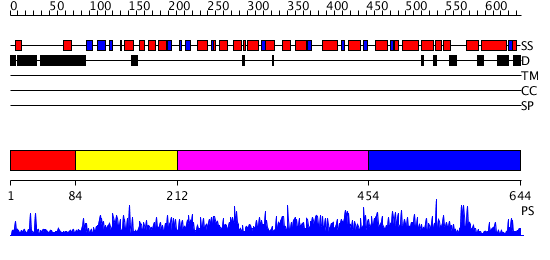
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..83] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [84..211] | 3.072977 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [212..453] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [454..644] | 3.072935 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)