













| General Information: |
|
| Name(s) found: |
NCBP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 848 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
response to abscisic acid stimulus
[IMP]
RNA splicing, via endonucleolytic cleavage and ligation [IMP] primary microRNA processing [IMP] long-day photoperiodism, flowering [IMP] translation [ISS] organ morphogenesis [NAS] |
| Molecular Function: |
RNA cap binding
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNWKTLLLR IGEKGPEYGT SSDYKDHIET CFGVIRREIE RSGDQVLPFL LQCAEQLPHK 60
61 IPLYGTLIGL LNLENEDFVQ KLVESVHANF QVALDSGNCN SIRILLRFMT SLLCSKVIQP 120
121 ASLIVVFETL LSSAATTVDE EKGNPSWQPQ ADFYVICILS SLPWGGSELA EQVPDEIERV 180
181 LVGIQAYLSI RKNSSTSGLN FFHNGEFESS LAEKDFVEDL LDRIQSLASN GWKLESVPRP 240
241 HLSFEAQLVA GKFHELRPIK CMEQPSPPSD HSRAYSGKQK HDALTRYPQR IRRLNIFPAN 300
301 KMEDVQPIDR FVVEEYLLDV LFYLNGCRKE CASYMANLPV TFRYEYLMAE TLFSQILLLP 360
361 QPPFKTLYYT LVIMDLCKAL PGAFPAVVAG AVRALFEKIS DLDMESRTRL ILWFSHHLSN 420
421 FQFIWPWEEW AFVLDLPKWA PKRVFVQEIL QREVRLSYWD KIKQSIENAT ALEELLPPKA 480
481 GPNFMYSLEE GKEKTEEQQL SAELSRKVKE KQTARDMIVW IEETIYPVHG FEVTLTIVVQ 540
541 TLLDIGSKSF THLVTVLERY GQVFSKLCPD NDKQVMLLSQ VSTYWKNNVQ MTAVAIDRMM 600
601 GYRLVSNQAI VRWVFSPENV DQFHVSDQPW EILGNALNKT YNRISDLRKD ISNITKNVLV 660
661 AEKASANARV ELEAAESKLS LVEGEPVLGE NPAKMKRLKS TVEKTGEAEL SLRESLEAKE 720
721 ALLNRALSET EVLLLLLFQS FLGVLKERLP DPTKVRSVQD LKSIGAEDDK PSAMDVDSEN 780
781 GNPKKSCEVG EREQWCLSTL GYLTAFTRQY ASEIWPHMEK LESEVFSGED VHPLFLQAIS 840
841 SALQFPLH |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
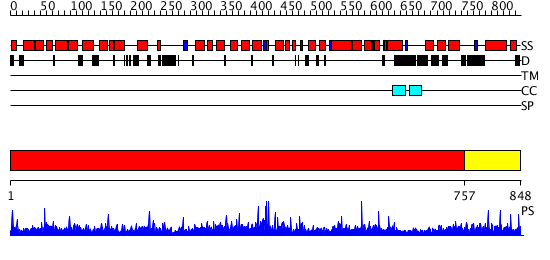
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..756] | 1000.0 | CBP80, 80KDa nuclear cap-binding protein | |
| 2 | View Details | [757..848] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)