













| General Information: |
|
| Name(s) found: |
FAB1C_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1648 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
cellular protein metabolic process
[IEA]
phosphatidylinositol metabolic process [IEA] |
| Molecular Function: |
ATP binding
[IEA]
phosphatidylinositol phosphate kinase activity [IEA] protein binding [IEA] 1-phosphatidylinositol-4-phosphate 5-kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGIPDGSLLD LIDKVRSWIT SDSSDSLFLL SSSKQDFGIM PIVSKMCHDC GTKVEQGYCC 60
61 LSCGSCWCKS CSDTEESKMK LCRECDAEVR ELRVKSYDKV HPRDSPDPPS SLATESESLA 120
121 SSLEIRDCRN MASIRCYPSR GEEEEARYCG KQLLSPSSDN YQDSSDIESG SVSARHELFS 180
181 CKSSAGSSPH DSPLRNNFSP LGRFVQHAKD LRSPTVCSFD NHQEQLLADN LVKPGQGVLE 240
241 QEDHEEEEDK LQQPLDFENN GRIWYPPPPE DENDDAESNY FHYDDEDDDI GDSATEFSLS 300
301 SSFSSHIPTK EKLGENSNEP LRTVVHDHFR ALVAELLRGE ELSPSDDGSA GEWLDIVTAL 360
361 AWQAANFVKP DTRAGGSMDP GNYVKIKCVA SGNQNESILI RGIVCSKNIT HKRMISQYKN 420
421 PRVMLLAGSL EYQRVAGQLA SFNTLLQQEN EHMKAIIAKI ESLRPNVLLV EKSASSYAQQ 480
481 YLLEKEISLV LNVKRSLLDR IARCTGAVLC PSLDSISTAR LGHCELFRTE RVLEQHEAGN 540
541 QSNRKPSRTL MYFEGCPRRL GCTVVLRGSC REELKKVKHV IQYAVFAAYH LSLETSFLAD 600
601 EGASLPKIRL KQPGMVRTAS QRRIIDEGIS LITQSPTETD SQALLETAAH EDEHTAPMPE 660
661 HEVCESLCED FDPTQIFPPS SEVETEQSDT LNGDFANNLV TRSYSSNQLN DLHEPTLCLS 720
721 SEIPETPTQQ PSGEEDNGRG EEENQLVNPQ DLPQHESFYE DDVSSEYFSA ADSHQSILVS 780
781 FSSRCVLKES VCERSRLLRI KFYGSFDKPL GRYLKDDLFD KTSSCRSCKE LVDAHVLCYS 840
841 HQNGNLTINV RRLPSMKLPG EQDGKIWMWH RCLRCAHVDG VPPATRRVVM SDAAWGLSFG 900
901 KFLELSFSNH ATANRVASCG HSLQRDCLRF YGFGNMVAFF RYSPINILTV LLPPSMLEFN 960
961 SHPQQEWIRT EAAELVGKMR TMYTEISDML NRMEEKSSLL EPEQSEACDL HSRIIGLIDQ1020
1021 LVKEKDEYDD ALQPIFEENL QIQGSLDILE LNRLRRALMI GAHAWDHQLY LLNSQLKKAS1080
1081 VFKTGDDNAP RNPEMHDPPK IDRRMQEGSD ERDEQSHTDS EANGDNKDPE NIPSPGTSLS1140
1141 ERIDSAWLGS FQNLEKAETI AETEGFSAVN SSLRRLARPI RVQSFDSAIR FQERIQKGLP1200
1201 PSSLYLSTLR SFHASGEYRN MVRDPVSNVM RTYSQMLPLE VQKLDLIVGS APTYISSASQ1260
1261 MADGARMLIP QRGLNDIVVP VYDDDPASVV SYAINSKEYK EWIVNKGLAS SSSSSNLNNR1320
1321 ESEPSAFSTW RSLSMDVDYI QHAVYGSSQD DRKSPHLTIS FSDRASSSST ATEGKVKFSV1380
1381 TCYFATQFDT LRKTCCPSEV DFVRSLSRCQ RWSAQGGKSN VYFAKSLDER FIIKQVVKTE1440
1441 LDSFEDFAPE YFKYLKESLS SGSPTCLAKI LGIYQVSIKH PKGGKETKMD LMVMENLFYN1500
1501 RRISRIYDLK GSARSRYNPN TSGADKVLLD MNLLETLRTE PIFLGSKAKR SLERAIWNDT1560
1561 NFLASVDVMD YSLLVGFDEE RKELVLGIID FMRQYTWDKH LETWVKASGI LGGPKNASPT1620
1621 IVSPKQYKRR FRKAMTTYFL TVPEPWTS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
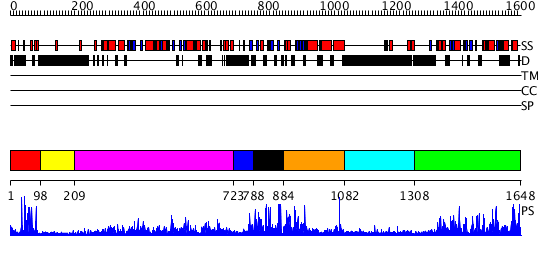
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..97] | 16.522879 | Hrs | |
| 2 | View Details | [98..208] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [209..722] | 119.0 | Crystal structure of the chaperonin from Thermococcus strain KS-1 (FormIII crystal complexed with ADP) | |
| 4 | View Details | [723..787] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [788..883] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [884..1081] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1082..1307] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1308..1648] | 77.30103 | Phosphatidylinositol phosphate kinase IIbeta, PIPK IIbeta |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)