













| General Information: |
|
| Name(s) found: |
AIB_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 566 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
response to abscisic acid stimulus
[IMP]
response to wounding [IEP] regulation of transcription [TAS] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [IDA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNMSDLGWDD EDKSVVSAVL GHLASDFLRA NSNSNQNLFL VMGTDDTLNK KLSSLVDWPN 60
61 SENFSWNYAI FWQQTMSRSG QQVLGWGDGC CREPNEEEES KVVRSYNFNN MGAEEETWQD 120
121 MRKRVLQKLH RLFGGSDEDN YALSLEKVTA TEIFFLASMY FFFNHGEGGP GRCYSSGKHV 180
181 WLSDAVNSES DYCFRSFMAK SAGIRTIVMV PTDAGVLELG SVWSLPENIG LVKSVQALFM 240
241 RRVTQPVMVT SNTNMTGGIH KLFGQDLSGA HAYPKKLEVR RNLDERFTPQ SWEGYNNNKG 300
301 PTFGYTPQRD DVKVLENVNM VVDNNNYKTQ IEFAGSSVAA SSNPSTNTQQ EKSESCTEKR 360
361 PVSLLAGAGI VSVVDEKRPR KRGRKPANGR EEPLNHVEAE RQRREKLNQR FYALRSVVPN 420
421 ISKMDKASLL GDAISYIKEL QEKVKIMEDE RVGTDKSLSE SNTITVEESP EVDIQAMNEE 480
481 VVVRVISPLD SHPASRIIQA MRNSNVSLME AKLSLAEDTM FHTFVIKSNN GSDPLTKEKL 540
541 IAAFYPETSS TQPPLPSSSS QVSGDI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
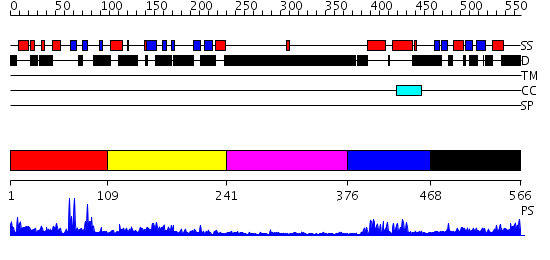
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..108] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [109..240] | 1.28 | Crystal structure of an hypothetical protein | |
| 3 | View Details | [241..375] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [376..467] | 14.39794 | SREBP-1a | |
| 5 | View Details | [468..566] | 0.968 | ACT domain protein from Streptococcus pneumoniae |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 |
|
||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)