













| General Information: |
|
| Name(s) found: |
PABP6_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 537 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
mRNA metabolic process
[TAS]
translational initiation [TAS] |
| Molecular Function: |
translation initiation factor activity
[ISS]
RNA binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALVKTETQA LGNHQHSSRF GSLYVGDLSP DVTEKDLIDK FSLNVPVVSV HLCRNSVTGK 60
61 SMCYAYINFD SPFSASNAMT RLNHSDLKGK AMRIMWSQRD LAYRRRTRTG FANLYVKNLD 120
121 SSITSSCLER MFCPFGSILS CKVVEENGQS KGFGFVQFDT EQSAVSARSA LHGSMVYGKK 180
181 LFVAKFINKD ERAAMAGNQD STNVYVKNLI ETVTDDCLHT LFSQYGTVSS VVVMRDGMGR 240
241 SRGFGFVNFC NPENAKKAME SLCGLQLGSK KLFVGKALKK DERREMLKQK FSDNFIAKPN 300
301 MRWSNLYVKN LSESMNETRL REIFGCYGQI VSAKVMCHEN GRSKGFGFVC FSNCEESKQA 360
361 KRYLNGFLVD GKPIVVRVAE RKEDRIKRLQ QYFQAQPRQY TQAPSAPSPA QPVLSYVSSS 420
421 YGCFQPFQVG TSYYYMGNQV PQMSGHQNIT TYVPAGKVPL KERRSMHLVY KHPAYPVAKR 480
481 GAKQTLVFKG EVNRNLEAAT CSKATTSEEN RKEERRLTLS GKLSPEVKVE ESGKQLQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
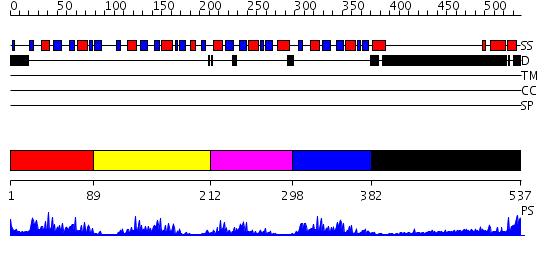
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | 48.69897 | Poly(A)-binding protein | |
| 2 | View Details | [89..211] | 9.045757 | NMR Structure of RRM2 from Human Polypyrimidine Tract Binding Protein Isoform 1 (PTB1) | |
| 3 | View Details | [212..297] | 3.69 | NMR Structure of the N-terminal RRM domain of Cleavage stimulation factor 64 KDa subunit | |
| 4 | View Details | [298..381] | 22.154902 | No description for 2d9pA was found. | |
| 5 | View Details | [382..537] | 3.30103 | Sec24 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)