













| General Information: |
|
| Name(s) found: |
TPS7_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 851 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
metabolic process
[IEA]
trehalose biosynthetic process [IEA][ISS] |
| Molecular Function: |
transferase activity, transferring glycosyl groups
[ISS]
trehalose-phosphatase activity [IGI][IMP][ISS] [NOT]alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity [IGI][IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MISRSYTNLL DLASGNFPVM GRERRRLPRV MTVPGNVSEF DEDQAYSVSS DNPSSVSSDR 60
61 MIIVANRLPL KAEKRNGSWS FSWDQDSLYL QLKDGLPEDM EILYVGSLSV DVDSNEQDDV 120
121 AQILLDKFKC VPTFFPPDLQ SKFYDGFCKR QIWPLFHYML PFSADHGGRF DRSLWEAYVA 180
181 TNKLFFQKVI EVINPDDDFV WIHDYHLMVL PTFLRRRFNR IRMGFFLHSP FPSSEIYRSL 240
241 PVREEILKAL LNSDLIGFHT FDYARHFLTC CSRMLGLEYQ SKRGYIGLEY YGRTVGIKIM 300
301 PVGINMGRIQ SVMRYSEEEG KVMELRNRFE GKTVLLGIDD MDIFKGINLK LLAMEQMLRQ 360
361 HPNWRGRAVL VQIVNPARGK GIDVEEIRGE IEESCRRING EFGKPGYQPI IYIDTPVSIN 420
421 EINAYYHIAE CVVVTAVRDG MNLTPYEYIV CRQGLLGSES DFSGPKKSML VASEFIGCSP 480
481 SLSGAIRVNP WNVEATGEAL NEALSMSDAE KQLRHEKHFR YVSTHDVAYW SRSFLQDLER 540
541 ICVDHFKKRC WGMGISFGFR VVALDPNFRK LSIPCIVSDY KRAKSRAILL DYDGTLMPQN 600
601 SINKAPSQEV LNFLDALCED KKNSIFIVSG RGRESLSKWF TPCKKIGIAA EHGYFLKWSG 660
661 SEEWETCGQS SDFGWMQIVE PVMKQYTEST DGSSIEIKES ALVWQYRDAD PGFGSLQAKE 720
721 MLEHLESVLA NEPVAVKSGH YIVEVKPQGV SKGSVSEKIF SSMAGKGKPV DFVLCIGDDR 780
781 SDEDMFEAIG NAMSKRLLCD NALVFACTVG QKPSKAKYYL DDTTEVTCML ESLAEASEAS 840
841 NFSMRELDEA L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
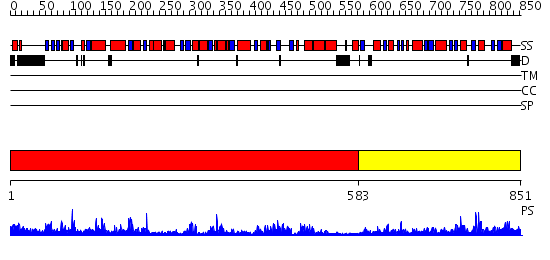
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..582] | 81.045757 | Trehalose-6-phosphate from E. coli bound with UDP-2-fluoro glucose. | |
| 2 | View Details | [583..851] | 37.0 | Crystal structure of NYSGRC target T1436: A Hypothetical protein yidA. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)