













| General Information: |
|
| Name(s) found: |
PLRX2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 847 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endomembrane system
[IEA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
protein binding
[ISS]
structural constituent of cell wall [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERPFGCFFI LLLISYTVVA TFDDEPSFPE NADLTKDLEQ KCFSINKVDP NLKFENDRLK 60
61 RAYIALQAWK KAIYSDPFKT TANWVGSDVC SYNGVYCAPA LDDDSLTVVA GVDLNHADIA 120
121 GHLPPELGLM TDLALFHINS NRFCGIIPKS LSKLALMYEF DVSNNRFVGQ FPEVSLSWPS 180
181 LKFLDLRYNE FEGSLPSEIF DKDLDAIFLN NNRFESVIPG TIGKSKASVV TFANNKFSGC 240
241 IPKSIGNMKN LNEIVFTGNN LTGCFPNEIG LLNNVTVFDA SKNGFVGSLP STLSGLASVE 300
301 QLDLSHNKLT GFVVDKFCKL PNLDSFKFSY NFFNGEAESC VPGRNNGKQF DDTNNCLQNR 360
361 PSQKPAKQCL PVVSRPVDCS KDKCSGGSNG GSSPSPNPPR TSEPKPSKPE PVMPKPSDSS 420
421 KPETPKTPEQ PSPKPQPPKH ESPKPEEPEN KHELPKQKES PKPQPSKPED SPKPEQPKPE 480
481 ESPKPEQPQI PEPTKPVSPP NEAQGPTPDD PYDASPVKNR RSPPPPKVED TRVPPPQPPM 540
541 PSPSPPSPIY SPPPPVHSPP PPVYSSPPPP HVYSPPPPVA SPPPPSPPPP VHSPPPPPVF 600
601 SPPPPVFSPP PPSPVYSPPP PSHSPPPPVY SPPPPTFSPP PTHNTNQPPM GAPTPTQAPT 660
661 PSSETTQVPT PSSESDQSQI LSPVQAPTPV QSSTPSSEPT QVPTPSSSES YQAPNLSPVQ 720
721 APTPVQAPTT SSETSQVPTP SSESNQSPSQ APTPILEPVH APTPNSKPVQ SPTPSSEPVS 780
781 SPEQSEEVEA PEPTPVNPSS VPSSSPSTDT SIPPPENNDD DDDGDFVLPP HIGFQYASPP 840
841 PPMFQGY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
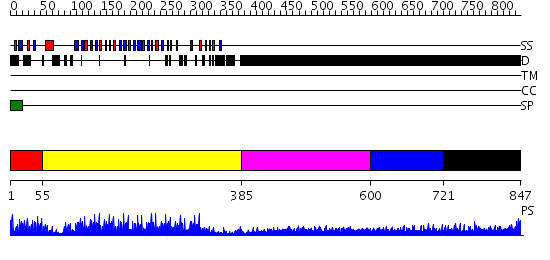
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..54] | 11.045757 | Rna1p (RanGAP1), N-terminal domain | |
| 2 | View Details | [55..384] | 48.522879 | Polygalacturonase inhibiting protein PGIP | |
| 3 | View Details | [385..599] | 1.354996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [600..720] | 4.107997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [721..847] | N/A | No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|