













| General Information: |
|
| Name(s) found: |
MAOP4_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 646 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
malate metabolic process
[ISS][TAS]
fatty acid biosynthetic process [TAS] |
| Molecular Function: |
zinc ion binding
[IDA]
malic enzyme activity [ISS] malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+) activity [IDA] oxidoreductase activity, acting on NADH or NADPH, NAD or NADP as acceptor [ISS] cobalt ion binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MISLTPSLFL NKTVVPGCST RLSLRQPRTI ITPPASLRVF SSLGSNQDPT GSVLIETTAT 60
61 SSSSLETSAA DIVPKSTVSG GVQDVYGEDA ATEDMPITPW SLSVASGYTL LRDPHHNKGL 120
121 AFSHRERDAH YLRGLLPPTV ISQDLQVKKI MHTLRQYQVP LQKYMAMMDL QETNERLFYK 180
181 LLIDHVEELL PVIYTPTVGE ACQKYGSIFL RPQGLFISLK EKGKIHEVLR NWPEKNIQVI 240
241 VVTDGERILG LGDLGCQGMG IPVGKLSLYT ALGGVRPSAC LPVTIDVGTN NEKLLNDEFY 300
301 IGLRQRRATG EEYSELMHEF MTAVKQNYGE KVVIQFEDFA NHNAFDLLAK YGTTHLVFND 360
361 DIQGTASVVL AGLIAALRFV GGSLSDHRFL FLGAGEAGTG IAELIALEIS KKSHIPLEEA 420
421 RKNIWLVDSK GLIVSSRKES IQHFKKPWAH DHEPIRELVD AVKAIKPTVL IGTSGVGQTF 480
481 TQDVVETMAK LNEKPIILSL SNPTSQSECT AEEAYTWSQG RAIFASGSPF APVEYEGKTF 540
541 VPGQANNAYI FPGFGLGLIM SGTIRVHDDM LLAASEALAE ELMEEHYEKG MIYPPFRNIR 600
601 KISARIAAKV AAKAYELGLA TRLPQPKELE QCAESSMYSP SYRSYR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
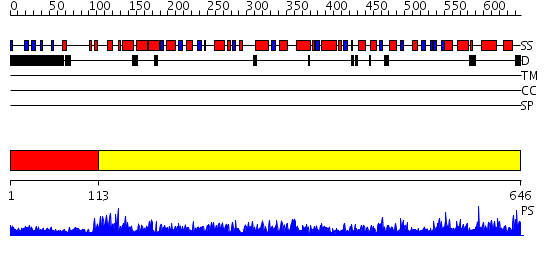
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..112] | 1.091986 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [113..646] | 1000.0 | Crystal structure of a human malic enzyme |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.74 |
Source: Reynolds et al. (2008)