













| General Information: |
|
| Name(s) found: |
CMT3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 839 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
chromatin [IEA] |
| Biological Process: |
negative regulation of gene expression, epigenetic
[IGI]
DNA methylation [IMP] chromatin silencing [IGI] DNA methylation on cytosine within a CNG sequence [IMP] histone H3-K9 methylation [IGI] zygote asymmetric cytokinesis in the embryo sac [IMP] |
| Molecular Function: |
DNA (cytosine-5-)-methyltransferase activity
[IMP]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPKRKRPAT KDDTTKSIPK PKKRAPKRAK TVKEEPVTVV EEGEKHVARF LDEPIPESEA 60
61 KSTWPDRYKP IEVQPPKASS RKKTKDDEKV EIIRARCHYR RAIVDERQIY ELNDDAYVQS 120
121 GEGKDPFICK IIEMFEGANG KLYFTARWFY RPSDTVMKEF EILIKKKRVF FSEIQDTNEL 180
181 GLLEKKLNIL MIPLNENTKE TIPATENCDF FCDMNYFLPY DTFEAIQQET MMAISESSTI 240
241 SSDTDIREGA AAISEIGECS QETEGHKKAT LLDLYSGCGA MSTGLCMGAQ LSGLNLVTKW 300
301 AVDMNAHACK SLQHNHPETN VRNMTAEDFL FLLKEWEKLC IHFSLRNSPN SEEYANLHGL 360
361 NNVEDNEDVS EESENEDDGE VFTVDKIVGI SFGVPKKLLK RGLYLKVRWL NYDDSHDTWE 420
421 PIEGLSNCRG KIEEFVKLGY KSGILPLPGG VDVVCGGPPC QGISGHNRFR NLLDPLEDQK 480
481 NKQLLVYMNI VEYLKPKFVL MENVVDMLKM AKGYLARFAV GRLLQMNYQV RNGMMAAGAY 540
541 GLAQFRLRFF LWGALPSEII PQFPLPTHDL VHRGNIVKEF QGNIVAYDEG HTVKLADKLL 600
601 LKDVISDLPA VANSEKRDEI TYDKDPTTPF QKFIRLRKDE ASGSQSKSKS KKHVLYDHHP 660
661 LNLNINDYER VCQVPKRKGA NFRDFPGVIV GPGNVVKLEE GKERVKLESG KTLVPDYALT 720
721 YVDGKSCKPF GRLWWDEIVP TVVTRAEPHN QVIIHPEQNR VLSIRENARL QGFPDDYKLF 780
781 GPPKQKYIQV GNAVAVPVAK ALGYALGTAF QGLAVGKDPL LTLPEGFAFM KPTLPSELA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
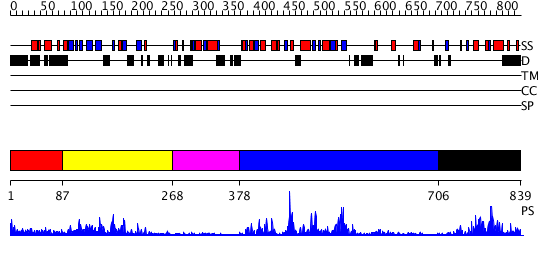
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [87..267] | 31.045757 | Crystal structure of the proximal BAH domain of polybromo | |
| 3 | View Details | [268..377] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [378..705] | 47.522879 | DNA methylase HaeIII | |
| 5 | View Details | [706..839] | 9.295993 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)