













| General Information: |
|
| Name(s) found: |
gi|7484958
[NCBI NR]
gi|7267482 [NCBI NR] gi|5731762 [NCBI NR] gi|15236488 [NCBI NR] |
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 707 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
plant-type cell wall organization
[IEA]
|
| Molecular Function: |
structural constituent of cell wall
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFMIKYNIKP KQRKKIVAMG VIRTSRTMGL GQCMVYVVVL SAIAATVTSY PYTSPQTPHY 60
61 NSPSHEHKIP KYTPHPKPSI YSSSPPPSYY SPSPKVDYKS PPPSYVYSSP PPPYYSPSPK 120
121 VDYKSLPPPY VYSSPPPPYY SPSPKVNYKS PPPPYVYSSP PPPYYSPSPK GDYKSPPPPY 180
181 VYSSPPPPYY SPSPKVDYKS PPPPYVYSSP PPPYYSPTPK VDYKSPPPPY VYSSPPPPYY 240
241 SPSPKVDYKS PPPPYVYSSP PPPYYSPSPK VNYKSPPPPY VYGSPPPPYY SPSPKVDYKS 300
301 PPPPYVYSSP PPPYYSPSPK VNYKSPPPPY VYGSPPPPYY SPSPKVDYKS PPPPYVYSSP 360
361 PPPYYSPSPK VDYKSPPPPY VYSSTPPPYY SPSPKVDYKS PPPPYVYSSP PPPYYSPSPK 420
421 VDYKSPPPPY IYSSTPLPYY SPSPKVDYKS PPPPYVYSSP PPPYYSPSPK VDYKPPPPPY 480
481 VYSSPPPPYY SPSPKVDYKS PPPPYVYSFP PPPYYSPSPK VDYKSPPPPY VYSSPPPPYY 540
541 SPSPKVNYKS PPPPYVYSSP PPPYYSPSPK VEYKSPPPPY IYSSPPPPYY APSPKVDYKS 600
601 PPPPYVYSSP PPPYYSPSPK VDYKSPPPPY VYSSPPPPYY SPSPKVNYKS PPPPYVYSSP 660
661 PPPYYSPSPK VDYKSSPPQY VYSSPPTPYY SPSPKVTYKS PPPPYVY |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
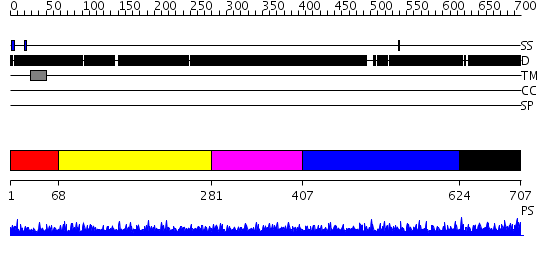
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..67] | 1.017995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [68..280] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [281..406] | 1.026992 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [407..623] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [624..707] | N/A | No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|