













| General Information: |
|
| Name(s) found: |
UBP17_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 731 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IEA]
|
| Biological Process: |
ubiquitin-dependent protein catabolic process
[IEA][ISS]
|
| Molecular Function: |
zinc ion binding
[IEA]
cysteine-type endopeptidase activity [ISS] ubiquitin thiolesterase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMLVFLLIRR QWRSASVRRE EVIRLIALAT EESYLAEEVR PATVDYGGDS VSDVYRCAVC 60
61 LYPTTTRCSQ CKSVRYCSSK CQILHWRRGH KEECRSPDYD EEKEEYVQSD YDAKESNVDF 120
121 PSRGTAYESS SNVSVDVACD MSTSRPSIHK VQPRSEAVDF TTSLNIKDNL YETRPLSRKK 180
181 SRNRTDKVES ASNYSKGKTD AKLRKLGNQN SRRSGDSANM SISDQFLSVG FEEEMNALKH 240
241 ERITSEPSSA SAAMSSSSTL LLPSKANSKP KVSQASSSGL KTSVQKVVQH FRPPQSSKKS 300
301 QPSSSIDEMS FSYELFVKLY CDRVELQPFG LVNLGNSCYA NAVLQCLAFT RPLISYLIRG 360
361 LHSKTCRKKS WCFVCEFEHL ILKARGGESP LSPIKILSKL QKIGKHLGPG KEEDAHEFLR 420
421 CAVDTMQSVF LKEAPAAGPF AEETTLVGLT FGGYLHSKIK CMACLHKSER PELMMDLTVE 480
481 IDGDIGSLEE ALAQFTAYEV LDGENRYFCG RCKSYQKAKK KLMILEGPNI LTVVLKRFQS 540
541 DNFGKLSKPI HFPELLDISP YMSDPNHGDH PVYSLYAVVV HLDAMSTLFS GHYVCYIKTL 600
601 DGDWFKIDDS NVFPVQLETV LLEGAYMLLY ARDSPRPVSK NGGRKSKQRR NLAAIPSRKG 660
661 NKKQRDGDNN SLLPRVDWSS GSLSSMFSSS DTTSSCSTKD SSGIENLSDY LFGGVEPVWK 720
721 WDRHNKSQTF D |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
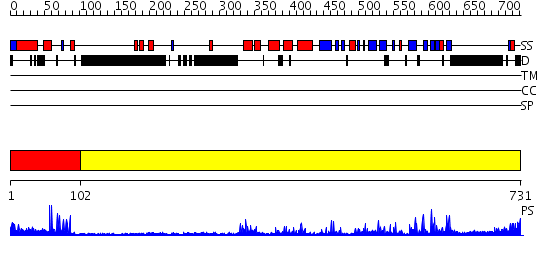
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] | 4.0 | No description for 2oddA was found. | |
| 2 | View Details | [102..731] | 55.0 | Crystal structure of HAUSP |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)