













| General Information: |
|
| Name(s) found: |
CAMK5_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 601 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
protein amino acid phosphorylation
[IEA]
|
| Molecular Function: |
ATP binding
[IEA]
calcium-dependent protein serine/threonine phosphatase activity [ISS] calcium ion binding [ISS] protein kinase activity [IEA] protein serine/threonine kinase activity [IEA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGLCTSKPNS SNSDQTPARN SPLPASESVK PSSSSVNGED QCVTTTNNEG KKSPFFPFYS 60
61 PSPAHYFFSK KTPARSPATN STNSTPKRFF KRPFPPPSPA KHIRAVLARR HGSVKPNSSA 120
121 IPEGSEAEGG GVGLDKSFGF SKSFASKYEL GDEVGRGHFG YTCAAKFKKG DNKGQQVAVK 180
181 VIPKAKMTTA IAIEDVRREV KILRALSGHN NLPHFYDAYE DHDNVYIVME LCEGGELLDR 240
241 ILSRGGKYTE EDAKTVMIQI LNVVAFCHLQ GVVHRDLKPE NFLFTSKEDT SQLKAIDFGL 300
301 SDYVRPDERL NDIVGSAYYV APEVLHRSYS TEADIWSVGV IVYILLCGSR PFWARTESGI 360
361 FRAVLKADPS FDDPPWPLLS SEARDFVKRL LNKDPRKRLT AAQALSHPWI KDSNDAKVPM 420
421 DILVFKLMRA YLRSSSLRKA ALRALSKTLT VDELFYLREQ FALLEPSKNG TISLENIKSA 480
481 LMKMATDAMK DSRIPEFLGQ LSALQYRRMD FEEFCAAALS VHQLEALDRW EQHARCAYEL 540
541 FEKEGNRPIM IDELASELGL GPSVPVHAVL HDWLRHTDGK LSFLGFVKLL HGVSSRTIKA 600
601 H |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
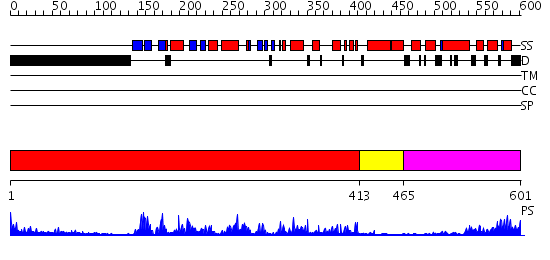
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..412] | 46.09691 | No description for 2psqA was found. | |
| 2 | View Details | [413..464] | 73.69897 | CALMODULIN-DEPENDENT PROTEIN KINASE FROM RAT | |
| 3 | View Details | [465..601] | 17.154902 | Troponin C |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)