













| General Information: |
|
| Name(s) found: |
UBQ9_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 322 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell wall
[IDA]
nucleolus [IDA] plasma membrane [IDA] intracellular [TAS] |
| Biological Process: |
protein modification process
[ISS]
ubiquitin-dependent protein catabolic process [ISS] |
| Molecular Function: |
protein binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSMQIHAKTL TEKTITIDVV SSDTINNVKA KIQDIEGIPL DQQRLIFSGK LLDDGRTLAD 60
61 YSIQKDSILH LALRLRGGMQ IFVKTLTGKT ITLEVESSDT IDNVKAKIQD KEGVPPDQQR 120
121 LIFAGKQLDD GRTLADYNIQ KESTLHLVLR LRGGMQIFVR TLTRKTIALE VESSDTTDNV 180
181 KAKIQDKEGI PPDQQRLIFA GKQLEDGRTL ADYNIQKEST LHLVLRLCGG MQIFVNTLTG 240
241 KTITLEVESS DTIDNVKAKI QDKERIQPDQ QRLIFAGEQL EDGYYTLADY NIQKESTLHL 300
301 VLRLRGGECF GFIFLFLLCF NS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
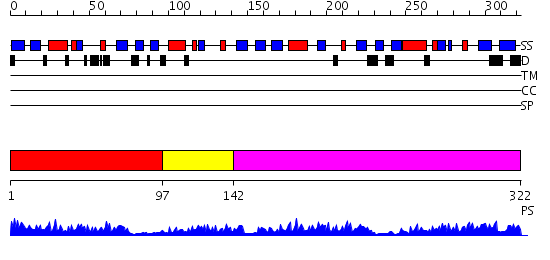
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | 30.221849 | Solution Structure of S5a UIM-1/Ubiquitin Complex | |
| 2 | View Details | [97..141] | 3.045757 | STRUCTURE DETERMINATION OF THE SMALL UBIQUITIN-RELATED MODIFIER SUMO-1, NMR, 10 STRUCTURES | |
| 3 | View Details | [142..322] | 20.221849 | Crystal Structure of ISG15, the Interferon-Induced Ubiquitin Cross Reactive Protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.66 |
Source: Reynolds et al. (2008)