













| General Information: |
|
| Name(s) found: |
WRK16_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1372 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
response to virus
[IMP]
plant-type hypersensitive response [IMP] |
| Molecular Function: |
transcription factor activity
[ISS]
protein binding [IPI][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTESEQIVYI SCIEEVRYSF VSHLSKALQR KGVNDVFIDS DDSLSNESQS MVERARVSVM 60
61 ILPGNRTVSL DKLVKVLDCQ KNKDQVVVPV LYGVRSSETE WLSALDSKGF SSVHHSRKEC 120
121 SDSQLVKETV RDVYEKLFYM ERIGIYSKLL EIEKMINKQP LDIRCVGIWG MPGIGKTTLA 180
181 KAVFDQMSGE FDAHCFIEDY TKAIQEKGVY CLLEEQFLKE NAGASGTVTK LSLLRDRLNN 240
241 KRVLVVLDDV RSPLVVESFL GGFDWFGPKS LIIITSKDKS VFRLCRVNQI YEVQGLNEKE 300
301 ALQLFSLCAS IDDMAEQNLH EVSMKVIKYA NGHPLALNLY GRELMGKKRP PEMEIAFLKL 360
361 KECPPAIFVD AIKSSYDTLN DREKNIFLDI ACFFQGENVD YVMQLLEGCG FFPHVGIDVL 420
421 VEKSLVTISE NRVRMHNLIQ DVGRQIINRE TRQTKRRSRL WEPCSIKYLL EDKEQNENEE 480
481 QKTTFERAQV PEEIEGMFLD TSNLSFDIKH VAFDNMLNLR LFKIYSSNPE VHHVNNFLKG 540
541 SLSSLPNVLR LLHWENYPLQ FLPQNFDPIH LVEINMPYSQ LKKLWGGTKD LEMLKTIRLC 600
601 HSQQLVDIDD LLKAQNLEVV DLQGCTRLQS FPATGQLLHL RVVNLSGCTE IKSFPEIPPN 660
661 IETLNLQGTG IIELPLSIVK PNYRELLNLL AEIPGLSGVS NLEQSDLKPL TSLMKISTSY 720
721 QNPGKLSCLE LNDCSRLRSL PNMVNLELLK ALDLSGCSEL ETIQGFPRNL KELYLVGTAV 780
781 RQVPQLPQSL EFFNAHGCVS LKSIRLDFKK LPVHYTFSNC FDLSPQVVND FLVQAMANVI 840
841 AKHIPRERHV TGFSQKTVQR SSRDSQQELN KTLAFSFCAP SHANQNSKLD LQPGSSSMTR 900
901 LDPSWRNTLV GFAMLVQVAF SEGYCDDTDF GISCVCKWKN KEGHSHRREI NLHCWALGKA 960
961 VERDHTFVFF DVNMRPDTDE GNDPDIWADL VVFEFFPVNK QRKPLNDSCT VTRCGVRLIT1020
1021 AVNCNTSIEN ISPVLSLDPM EVSGNEDEEV LRVRYAGLQE IYKALFLYIA GLFNDEDVGL1080
1081 VAPLIANIID MDVSYGLKVL AYRSLIRVSS NGEIVMHYLL RQMGKEILHT ESKKTDKLVD1140
1141 NIQSSMIATK EIEITRSKSR RKNNKEKRVV CVVDRGSRSS DLWVWRKYGQ KPIKSSPYPR1200
1201 SYYRCASSKG CFARKQVERS RTDPNVSVIT YISEHNHPFP TLRNTLAGST RSSSSKCSDV1260
1261 TTSASSTVSQ DKEGPDKSHL PSSPASPPYA AMVVKEEDME QWDNMEFDVD VEEDTFIPEL1320
1321 FPEDTFADMD KLEENSQTMF LSRRSSGGNM EAQGKNSSDD REVNLPSKIL NR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
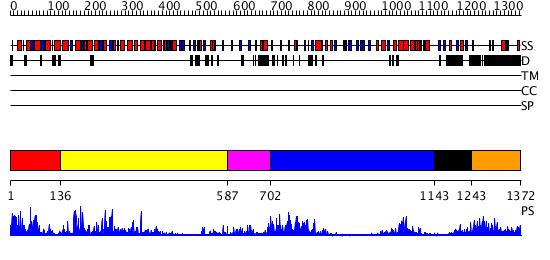
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..135] | 5.81899 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [136..586] | 11.522879 | Structure of the apoptotic protease-activating factor 1 bound to ADP | |
| 3 | View Details | [587..701] | 4.09691 | No description for 2v70A was found. | |
| 4 | View Details | [702..1142] | 7.0 | No description for 2z7xB was found. | |
| 5 | View Details | [1143..1242] | 6.154902 | No description for 2aydA was found. | |
| 6 | View Details | [1243..1372] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)