













| General Information: |
|
| Name(s) found: |
TOP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 916 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chromosome
[IEA]
|
| Biological Process: |
regulation of meristem growth
[IMP]
chromatin remodeling [IGI] stem cell maintenance [IGI] embryonic development ending in seed dormancy [IMP] post-embryonic development [IMP] shoot morphogenesis [IMP] organ formation [IMP] DNA topological change [ISS] DNA metabolic process [ISS] meristem structural organization [IMP] leaf morphogenesis [IMP] flower morphogenesis [IMP] |
| Molecular Function: |
protein binding
[IPI]
DNA topoisomerase type I activity [IGI][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGTETVSKPV MDNGSGDSDD DKPLAFKRNN TVASNSNQSK SNSQRSKAVP TTKVSPMRSP 60
61 VTSPNGTTPS NKTSIVKSSM PSSSSKASPA KSPLRNDMPS TVKDRSQLQK DQSECKIEHE 120
121 DSEDDRPLSS ILSGNKGPTS SRQVSSPQPE KKNNGDRPLD RASRIIKDES DDETPISSMF 180
181 RKKIDSGMSG GNQLSNDEKK PLVQKLHQNG STVKNEVPNG KVLGKRPLEK NSSADQSSLK 240
241 KAKISASPTS VKMKQDSVKK EIDDKGRVLV SPKMKAKQLS TREDGTDDDD DDDVPISKRF 300
301 KSDSSNSNTS SAKPKAVKLN STSSAAKPKA RNVVSPRSRA MTKNTKKVTK DSKYSTSSKS 360
361 SPSSGDGQKK WTTLVHNGVI FPPPYKPHGI KILYKGKPVD LTIEQEEVAT MFAVMRETDY 420
421 YTKPQFRENF WNDWRRLLGK KHVIQKLDDC DFTPIYEWHL EEKEKKKQMS TEEKKALKEE 480
481 KMKQEEKYMW AVVDGVKEKI GNFRVEPPGL FRGRGEHPKM GKLKKRIHPC EITLNIGKGA 540
541 PIPECPIAGE RWKEVKHDNT VTWLAFWADP INPKEFKYVF LGAGSSLKGL SDKEKYEKAR 600
601 NLTDHIDNIR TTYTKNFTAK DVKMRQIAVA TYLIDKLALR AGNEKDDDEA DTVGCCTLKV 660
661 GNVECIPPNK IKFDFLGKDS IQYVNTVEVE PLVYKAIGQF QAGKSKTDDL FDELDTSKLN 720
721 AHLKELVPGL TAKVFRTYNA SITLDEMLSQ ETKDGDVTQK IVVYQKANKE VAIICNHQRT 780
781 VSKTHGAQIE KLTARIEELK EVLKELKTNL DRAKKGKPPL EGSDGKKIRS LEPNAWEKKI 840
841 AQQSAKIEKM ERDMHTKEDL KTVALGTSKI NYLDPRITVA WCKRHEVPIE KIFTKSLLEK 900
901 FAWAMDVEPE YRFSRR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
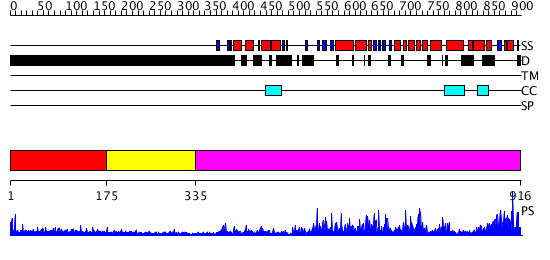
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..174] | 4.327997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [175..334] | 3.327996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [335..916] | 1000.0 | Eukaryotic DNA topoisomerase I, catalytic core; Eukaryotic DNA topoisomerase I, N-terminal DNA-binding fragment |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)