













| General Information: |
|
| Name(s) found: |
SUVH8_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 755 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
regulation of gene expression, epigenetic
[TAS]
|
| Molecular Function: |
histone methyltransferase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVSTPPTLLM LFDDGDAGPS TGLVHREKSD AVNEEAHATS VPPHAPPQTL WLLDNFNIED 60
61 SYDRDAGPST GPVHRERSDA VNEEAHATSI PPHAPPQTLW LLDNFNIEDS YDRDAGPSTS 120
121 PIDREASHEV NEDAHATSAP PHVMVSPLQN RRPFDQFNNQ PYDASAGPST GPGKRGRGRP 180
181 KGSKNGSRKP KKPKAYDNNS TDASAGPSSG LGKRRCGRPK GLKNRSRKPK KPKADDPNSK 240
241 MVISCPDFDS RITEAERESG NQEIVDSILM RFDAVRRRLC QLNYRKDKIL TASTNCMNLG 300
301 VRTNMTRRIG PIPGVQVGDI FYYWCEMCLV GLHRNTAGGI DSLLAKESGV DGPAATSVVT 360
361 SGKYDNETED LETLIYSGHG GKPCDQVLQR GNRALEASVR RRNEVRVIRG ELYNNEKVYI 420
421 YDGLYLVSDC WQVTGKSGFK EYRFKLLRKP GQPPGYAIWK LVENLRNHEL IDPRQGFILG 480
481 DLSFGEEGLR VPLVNEVDEE DKTIPDDFDY IRSQCYSGMT NDVNVDSQSL VQSYIHQNCT 540
541 CILKNCGQLP YHDNILVCRK PLIYECGGSC PTRMVETGLK LHLEVFKTSN CGWGLRSWDP 600
601 IRAGTFICEF TGVSKTKEEV EEDDDYLFDT SRIYHSFRWN YEPELLCEDA CEQVSEDANL 660
661 PTQVLISAKE KGNVGRFMNH NCWPNVFWQP IEYDDNNGHI YVRIGLFAMK HIPPMTELTY 720
721 DYGISCVEKT GEDEVIYKGK KICLCGSVKC RGSFG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
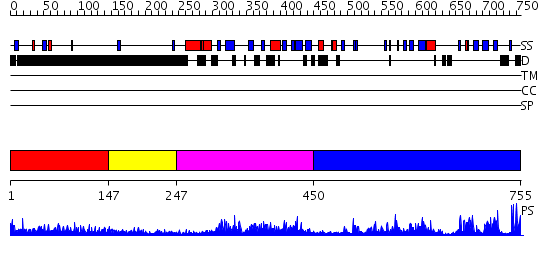
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..146] | 3.226997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [147..246] | 4.233992 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [247..449] | 36.853872 | No description for PF02182.8 was found. No confident structure predictions are available. | |
| 4 | View Details | [450..755] | 72.69897 | No description for 2r3aA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)