













| General Information: |
|
| Name(s) found: |
SEUSS_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 877 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
embryonic development
[IMP]
regulation of flower development [IGI] gynoecium development [IMP] multicellular organismal development [IMP] ovule development [IMP] |
| Molecular Function: |
DNA binding
[IDA]
protein binding [IPI][NAS] transcription cofactor activity [IMP] protein heterodimerization activity [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVPSEPPNPV GGGENVPPSI LGGQGGAPLP SQPAFPSLVS PRTQFGNNMS MSMLGNAPNI 60
61 SSLLNNQSFV NGIPGSMISM DTSGAESDPM SNVGFSGLSS FNASSMVSPR SSGQVQGQQF 120
121 SNVSANQLLA EQQRNKKMET QSFQHGQQQS MQQQFSTVRG GGLAGVGPVK MEPGQVSNDQ 180
181 QHGQVQQQQQ KMLRNLGSVK LEPQQIQAMR NLAQVKMEPQ HSEQSLFLQQ QQRQQQQQQQ 240
241 QQFLQMPGQS PQAQMNIFQQ QRLMQLQQQQ LLKSMPQQRP QLPQQFQQQN LPLRPPLKPV 300
301 YEPGMGAQRL TQYMYRQQHR PEDNNIEFWR KFVAEYFAPN AKKRWCVSMY GSGRQTTGVF 360
361 PQDVWHCEIC NRKPGRGFEA TAEVLPRLFK IKYESGTLEE LLYVDMPRES QNSSGQIVLE 420
421 YAKATQESVF EHLRVVRDGQ LRIVFSPDLK IFSWEFCARR HEELIPRRLL IPQVSQLGSA 480
481 AQKYQQAAQN ATTDSALPEL QNNCNMFVAS ARQLAKALEV PLVNDLGYTK RYVRCLQISE 540
541 VVNSMKDLID YSRETRTGPI ESLAKFPRRT GPSSALPGPS PQQASDQLRQ QQQQQQQQQQ 600
601 QQQQQQQQQQ QQQTVSQNTN SDQSSRQVAL MQGNPSNGVN YAFNAASAST STSSIAGLIH 660
661 QNSMKGRHQN AAYNPPNSPY GGNSVQMQSP SSSGTMVPSS SQQQHNLPTF QSPTSSSNNN 720
721 NPSQNGIPSV NHMGSTNSPA MQQAGEVDGN ESSSVQKILN EILMNNQAHN NSSGGSMVGH 780
781 GSFGNDGKGQ ANVNSSGVLL MNGQVNNNNN TNIGGAGGFG GGIGQSMAAN GINNINGNNS 840
841 LMNGRVGMMV RDPNGQQDLG NQLLGAVNGF NNFDWNA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
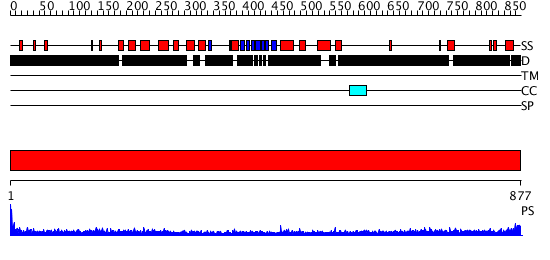
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..877] | 5.0 | No description for 1y0fB was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)