













| General Information: |
|
| Name(s) found: |
SCC12_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 810 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
nuclear cohesin complex [ISS] |
| Biological Process: |
mitosis
[IEP]
double-strand break repair [IMP] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFYSHCLVSR KGPLGAIWVA AYFFKKLKKS QVKATHIPSS VDQILQKELD ALTYRVLAYL 60
61 LLGVVRIYSK KVDFLFDDCN KALIGVKEFV AKERNREKTG VSLPASIECF SIALPERFEL 120
121 DAFDLGVLED FHGGNVKPHE DITLKDGSQE TERMDMYSME RFDMEEDLLF TFHETFSTNH 180
181 NENKHESFAH DMELDAENVR DTTEEASVRV VEAEPLDSNE PSRDHQNASR HREDPESDDI 240
241 LLEPQMSEDI RIAQEEDTVR ETICTIVQRL VDSHESSGDN LHRDGHTENL ESEKTSKKTS 300
301 CEEMQHDRSL PSECGIPEAI HGIEDQPSGA TRINGEKEIP EMSTLEKPEP VSVTGSRDLQ 360
361 EGVEKCRDHN EAEMADFELF HGSHKEQSET SEVNLHGSEK GFLSDMTVSK DPSSEFNATD 420
421 TPVTVTPKTP SRLKISEGGT SPQFSIIPTP AAKESSRVSR KRKCLIDDEV IIPNKVMKEM 480
481 IEDSSKLLAK RRNVPHTDCP ERRTKRFANP FRSFLEPLIQ YGSSDLQSLF CQPIKLKNWA 540
541 TTGTPKDTKI ARHKEKSSLD TVRSPGVILS SDQTENTQEI METPQAAALA GLKVTAGNSN 600
601 VVSVEMGASS TTSGTAHQTE NAAETPVKPS VIAPETPVRT SEQTVIAPET PVVSEQVEIA 660
661 PETPVRESMS KRFFKDPGTC YKKSRPASPF TSFEEHPSVY YVENRDLDTI LMNDEQVNAD 720
721 ERQDLQQETW SSRTRNVAKF LEKTFLEQRE REEEEKVSLL QLCRGRTQKE SARLFYETLV 780
781 LKTKGYVEVK QNHPYSDVFL MRVSRPQKAC |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
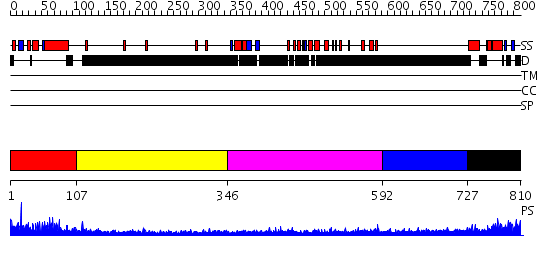
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | 52.387216 | No description for PF04825.4 was found. No confident structure predictions are available. | |
| 2 | View Details | [107..345] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [346..591] | 4.259996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [592..726] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [727..810] | 3.522879 | Sc Smc1hd:Scc1-C complex, ATPgS |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. | |||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)