













| General Information: |
|
| Name(s) found: |
RBCMT_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 482 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] |
| Biological Process: | NONE FOUND |
| Molecular Function: |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase activity
[IEA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSASVAVVSG FLRIPSIQKS QNPSFLFSRP KKSLVRPISA SSSELPENVR NFWKWLRDQG 60
61 VVSGKSVAEP AVVPEGLGLV ARRDIGRNEV VLEIPKRLWI NPETVTASKI GPLCGGLKPW 120
121 VSVALFLIRE KYEEESSWRV YLDMLPQSTD STVFWSEEEL AELKGTQLLS TTLGVKEYVE 180
181 NEFLKLEQEI LLPNKDLFSS RITLDDFIWA FGILKSRAFS RLRGQNLVLI PLADLINHNP 240
241 AIKTEDYAYE IKGAGLFSRD LLFSLKSPVY VKAGEQVYIQ YDLNKSNAEL ALDYGFVESN 300
301 PKRNSYTLTI EIPESDPFFG DKLDIAESNK MGETGYFDIV DGQTLPAGML QYLRLVALGG 360
361 PDAFLLESIF NNTIWGHLEL PVSRTNEELI CRVVRDACKS ALSGFDTTIE EDEKLLDKGK 420
421 LEPRLEMALK IRIGEKRVLQ QIDQIFKDRE LELDILEYYQ ERRLKDLGLV GEQGDIIFWE 480
481 TK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
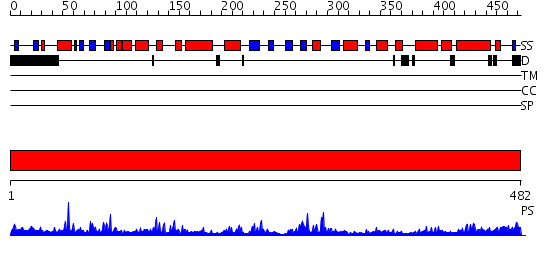
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..482] | 96.0 | RuBisCo LSMT C-terminal, substrate-binding domain; RuBisCo LSMT catalytic domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)