













| General Information: |
|
| Name(s) found: |
PLDD1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 868 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IDA]
vacuole [IDA] plasma membrane [IDA] |
| Biological Process: |
response to cold
[IEP][IMP]
phosphatidic acid metabolic process [IMP] programmed cell death [IMP] |
| Molecular Function: |
phospholipase D activity
[IDA][IMP][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEKVSEDVM LLHGDLDLKI VKARRLPNMD MFSEHLRRLF TACNACARPT DTDDVDPRDK 60
61 GEFGDKNIRS HRKVITSDPY VTVVVPQATL ARTRVLKNSQ EPLWDEKFNI SIAHPFAYLE 120
121 FQVKDDDVFG AQIIGTAKIP VRDIASGERI SGWFPVLGAS GKPPKAETAI FIDMKFTPFD 180
181 QIHSYRCGIA GDPERRGVRR TYFPVRKGSQ VRLYQDAHVM DGTLPAIGLD NGKVYEHGKC 240
241 WEDICYAISE AHHMIYIVGW SIFHKIKLVR ETKVPRDKDM TLGELLKYKS QEGVRVLLLV 300
301 WDDKTSHDKF GIKTPGVMGT HDEETRKFFK HSSVICVLSP RYASSKLGLF KQQASPSSSI 360
361 YIMTVVGTLF THHQKCVLVD TQAVGNNRKV TAFIGGLDLC DGRYDTPEHR ILHDLDTVFK 420
421 DDFHNPTFPA GTKAPRQPWH DLHCRIDGPA AYDVLINFEQ RWRKATRWKE FSLRLKGKTH 480
481 WQDDALIRIG RISWILSPVF KFLKDGTSII PEDDPCVWVS KEDDPENWHV QIFRSIDSGS 540
541 VKGFPKYEDE AEAQHLECAK RLVVDKSIQT AYIQTIRSAQ HFIYIENQYF LGSSYAWPSY 600
601 RDAGADNLIP MELALKIVSK IRAKERFAVY VVIPLWPEGD PKSGPVQEIL YWQSQTMQMM 660
661 YDVIAKELKA VQSDAHPLDY LNFYCLGKRE QLPDDMPATN GSVVSDSYNF QRFMIYVHAK 720
721 GMIVDDEYVL MGSANINQRS MAGTKDTEIA MGAYQPNHTW AHKGRHPRGQ VYGYRMSLWA 780
781 EHLGKTGDEF VEPSDLECLK KVNTISEENW KRFIDPKFSE LQGHLIKYPL QVDVDGKVSP 840
841 LPDYETFPDV GGKIIGAHSM ALPDTLTT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
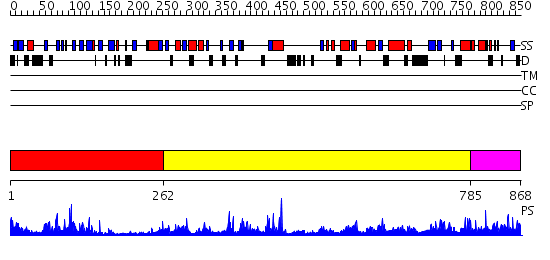
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..261] | 28.09691 | Synaptogamin I | |
| 2 | View Details | [262..784] | 42.30103 | Tungstate-inhibited phospholipase D from Streptomyces sp. strain PMF. | |
| 3 | View Details | [785..868] | 1.107985 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)