













| General Information: |
|
| Name(s) found: |
MSH7_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1109 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
mismatch repair
[IEA][ISS]
|
| Molecular Function: |
ATP binding
[IEA][ISS]
mismatched DNA binding [IEA] damaged DNA binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQRQRSILSF FQKPTAATTK GLVSGDAASG GGGSGGPRFN VKEGDAKGDA SVRFAVSKSV 60
61 DEVRGTDTPP EKVPRRVLPS GFKPAESAGD ASSLFSNIMH KFVKVDDRDC SGERSREDVV 120
121 PLNDSSLCMK ANDVIPQFRS NNGKTQERNH AFSFSGRAEL RSVEDIGVDG DVPGPETPGM 180
181 RPRASRLKRV LEDEMTFKED KVPVLDSNKR LKMLQDPVCG EKKEVNEGTK FEWLESSRIR 240
241 DANRRRPDDP LYDRKTLHIP PDVFKKMSAS QKQYWSVKSE YMDIVLFFKV GKFYELYELD 300
301 AELGHKELDW KMTMSGVGKC RQVGISESGI DEAVQKLLAR GYKVGRIEQL ETSDQAKARG 360
361 ANTIIPRKLV QVLTPSTASE GNIGPDAVHL LAIKEIKMEL QKCSTVYGFA FVDCAALRFW 420
421 VGSISDDASC AALGALLMQV SPKEVLYDSK GLSREAQKAL RKYTLTGSTA VQLAPVPQVM 480
481 GDTDAAGVRN IIESNGYFKG SSESWNCAVD GLNECDVALS ALGELINHLS RLKLEDVLKH 540
541 GDIFPYQVYR GCLRIDGQTM VNLEIFNNSC DGGPSGTLYK YLDNCVSPTG KRLLRNWICH 600
601 PLKDVESINK RLDVVEEFTA NSESMQITGQ YLHKLPDLER LLGRIKSSVR SSASVLPALL 660
661 GKKVLKQRVK AFGQIVKGFR SGIDLLLALQ KESNMMSLLY KLCKLPILVG KSGLELFLSQ 720
721 FEAAIDSDFP NYQNQDVTDE NAETLTILIE LFIERATQWS EVIHTISCLD VLRSFAIAAS 780
781 LSAGSMARPV IFPESEATDQ NQKTKGPILK IQGLWHPFAV AADGQLPVPN DILLGEARRS 840
841 SGSIHPRSLL LTGPNMGGKS TLLRATCLAV IFAQLGCYVP CESCEISLVD TIFTRLGASD 900
901 RIMTGESTFL VECTETASVL QNATQDSLVI LDELGRGTST FDGYAIAYSV FRHLVEKVQC 960
961 RMLFATHYHP LTKEFASHPR VTSKHMACAF KSRSDYQPRG CDQDLVFLYR LTEGACPESY1020
1021 GLQVALMAGI PNQVVETASG AAQAMKRSIG ENFKSSELRS EFSSLHEDWL KSLVGISRVA1080
1081 HNNAPIGEDD YDTLFCLWHE IKSSYCVPK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
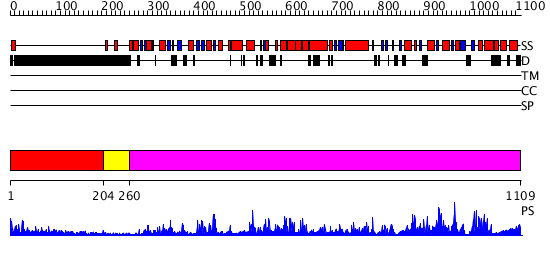
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..203] | 10.270997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [204..259] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [260..1109] | 1000.0 | Crystal Structure of E. coli DNA Mismatch Repair enzyme MutS, E38T mutant, in complex with a G.T mismatch |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.68 |
Source: Reynolds et al. (2008)