













| General Information: |
|
| Name(s) found: |
MEDEA_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 689 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
regulation of gene expression by genetic imprinting
[IMP]
negative regulation of transcription [IMP] seed morphogenesis [IGI] response to absence of light [IEP] |
| Molecular Function: |
transcription factor activity
[ISS]
sequence-specific DNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKENHEDDG EGLPPELNQI KEQIEKERFL HIKRKFELRY IPSVATHASH HQSFDLNQPA 60
61 AEDDNGGDNK SLLSRMQNPL RHFSASSDYN SYEDQGYVLD EDQDYALEED VPLFLDEDVP 120
121 LLPSVKLPIV EKLPRSITWV FTKSSQLMAE SDSVIGKRQI YYLNGEALEL SSEEDEEDEE 180
181 EDEEEIKKEK CEFSEDVDRF IWTVGQDYGL DDLVVRRALA KYLEVDVSDI LERYNELKLK 240
241 NDGTAGEASD LTSKTITTAF QDFADRRHCR RCMIFDCHMH EKYEPESRSS EDKSSLFEDE 300
301 DRQPCSEHCY LKVRSVTEAD HVMDNDNSIS NKIVVSDPNN TMWTPVEKDL YLKGIEIFGR 360
361 NSCDVALNIL RGLKTCLEIY NYMREQDQCT MSLDLNKTTQ RHNQVTKKVS RKSSRSVRKK 420
421 SRLRKYARYP PALKKTTSGE AKFYKHYTPC TCKSKCGQQC PCLTHENCCE KYCGCSKDCN 480
481 NRFGGCNCAI GQCTNRQCPC FAANRECDPD LCRSCPLSCG DGTLGETPVQ IQCKNMQFLL 540
541 QTNKKILIGK SDVHGWGAFT WDSLKKNEYL GEYTGELITH DEANERGRIE DRIGSSYLFT 600
601 LNDQLEIDAR RKGNEFKFLN HSARPNCYAK LMIVRGDQRI GLFAERAIEE GEELFFDYCY 660
661 GPEHADWSRG REPRKTGASK RSKEARPAR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
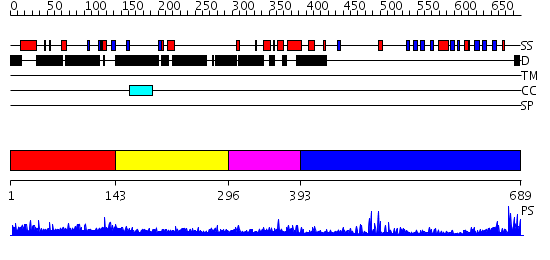
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..142] | 2.036984 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [143..295] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [296..392] | 0.989 | Solution Structure of the SMRT Deacetylase Activation Domain | |
| 4 | View Details | [393..689] | 31.09691 | No description for 2r3aA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)