













| General Information: |
|
| Name(s) found: |
HIS2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 281 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
histidine biosynthetic process
[TAS]
|
| Molecular Function: |
phosphoribosyl-ATP diphosphatase activity
[IDA]
phosphoribosyl-AMP cyclohydrolase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAVSYNALAQ SLARSSCFIP KPYSFRDTKL RSRSNVVFAC NDNKNIALQA KVDNLLDRIK 60
61 WDDKGLAVAI AQNVDTGAVL MQGFVNREAL STTISSRKAT FFSRSRSTLW TKGETSNNFI 120
121 NILDVYVDCD RDSIIYLGTP DGPTCHTGEE TCYYTSVFDQ LNNDEASGNK LALTTLYSLE 180
181 SIISKRKEES TVPQEGKPSW TRRLLTDDAL LCSKIREEAD ELCRTLEDNE EVSRTPSEMA 240
241 DVLYHAMVLL SKRGVKMEDV LEVLRKRFSQ SGIEEKQNRT K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
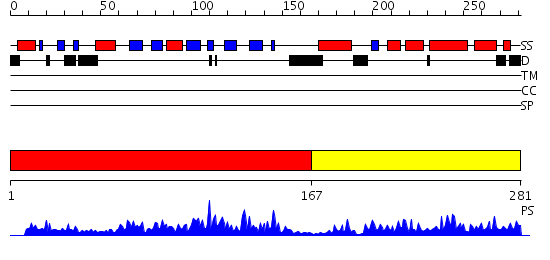
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..166] | 46.221849 | Crystal structure of Methanobacterium thermoautotrophicum phosphoribosyl-AMP cyclohydrolase HisI | |
| 2 | View Details | [167..281] | 27.69897 | Crystal Structure of Phosphoribosyl-ATP Pyrophosphatase from Chromobacterium violaceum (ATCC 12472). NESG TARGET CVR7 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)