













| General Information: |
|
| Name(s) found: |
HEM21_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 430 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] |
| Biological Process: |
porphyrin biosynthetic process
[TAS]
|
| Molecular Function: |
porphobilinogen synthase activity
[IEA][ISS]
metal ion binding [IEA] catalytic activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATTPIFNAS CSFPSTRGID CKSYIGLRSN VSKVSVASSR IATSQRRNLV VRASESGNGH 60
61 AKKLGMSDAE CEAAVAAGNV PEAPPVPPKP AAPVGTPIIK PLNLSRRPRR NRASPVTRAA 120
121 FQETDISPAN FVYPLFIHEG EEDTPIGAMP GCYRLGWRHG LVQEVAKARA VGVNSIVLFP 180
181 KVPEALKNST GDEAYNDNGL VPRTIRLLKD KYPDLIIYTD VALDPYSSDG HDGIVREDGV 240
241 IMNDETVHQL CKQAVSQARA GADVVSPSDM MDGRVGAIRS ALDAEGFQNV SIMSYTAKYA 300
301 SSFYGPFREA LDSNPRFGDK KTYQMNPANY REALIEARED EAEGADILLV KPGLPYLDII 360
361 RLLRDKSPLP IAAYQVSGEY SMIKAGGVLK MIDEEKVMME SLMCLRRAGA DIILTYFALQ 420
421 AATCLCGEKR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
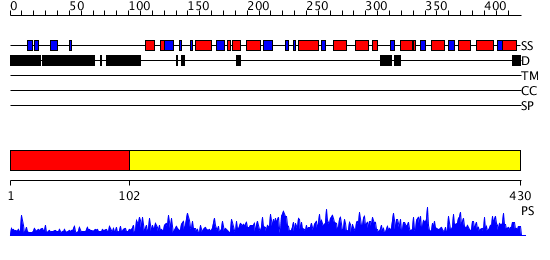
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [102..430] | 148.0 | Structure of the plant like 5-Aminolaevulinic Acid Dehydratase from Chlorobium vibrioforme |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)