













| General Information: |
|
| Name(s) found: |
HAT5_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 272 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA][ISS]
|
| Biological Process: |
response to salt stress
[IEP]
regulation of transcription, DNA-dependent [ISS] positive regulation of transcription [IDA] response to blue light [IEP] leaf morphogenesis [IMP] |
| Molecular Function: |
DNA binding
[IDA]
transcription factor activity [ISS] protein homodimerization activity [IDA] sequence-specific DNA binding [IDA] transcription activator activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESNSFFFDP SASHGNSMFF LGNLNPVVQG GGARSMMNME ETSKRRPFFS SPEDLYDDDF 60
61 YDDQLPEKKR RLTTEQVHLL EKSFETENKL EPERKTQLAK KLGLQPRQVA VWFQNRRARW 120
121 KTKQLERDYD LLKSTYDQLL SNYDSIVMDN DKLRSEVTSL TEKLQGKQET ANEPPGQVPE 180
181 PNQLDPVYIN AAAIKTEDRL SSGSVGSAVL DDDAPQLLDS CDSYFPSIVP IQDNSNASDH 240
241 DNDRSCFADV FVPTTSPSHD HHGESLAFWG WP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
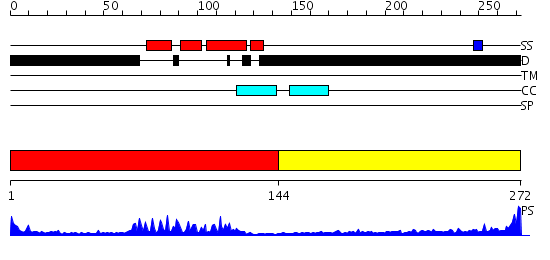
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..143] | 24.09691 | Crystal Structure of HoxA9 and Pbx1 homeodomains bound to DNA | |
| 2 | View Details | [144..272] | 15.825997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)