













| General Information: |
|
| Name(s) found: |
GLTB1_ARATH
[Swiss-Prot]
GLS1_ARATH [Swiss-Prot] |
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1648 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] membrane [IDA] apoplast [IDA] mitochondrion [IDA] chloroplast envelope [IDA] |
| Biological Process: |
photorespiration
[IMP][TAS]
positive regulation of glycine hydroxymethyltransferase activity [IDA] response to light stimulus [IEP] |
| Molecular Function: |
protein binding
[IPI]
glutamate synthase (ferredoxin) activity [IDA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAMQSLSPVP KLLSTTPSSV LSSDKNFFFV DFVGLYCKSK RTRRRLRGDS SSSSRSSSSL 60
61 SRLSSVRAVI DLERVHGVSE KDLSSPSALR PQVRFFTDIN FTNTQRAKFH PLWGSFKQVA 120
121 NLEDILSERG ACGVGFIANL DNIPSHGVVK DALIALGCME HRGGCGADND SGDGSGLMSS 180
181 IPWDFFNVWA KEQSLAPFDK LHTGVGMIFL PQDDTFMQEA KQVIENIFEK EGLQVLGWRE 240
241 VPVNVPIVGK NARETMPNIQ QVFVKIAKED STDDIERELY ICRKLIERAV ATESWGTELY 300
301 FCSLSNQTIV YKGMLRSEAL GLFYLDLQNE LYESPFAIYH RRYSTNTSPR WPLAQPMRFL 360
361 GHNGEINTIQ GNLNWMQSRE ASLKAAVWNG RENEIRPFGN PRGSDSANLD SAAEIMIRSG 420
421 RTPEEALMIL VPEAYKNHPT LSVKYPEVVD FYDYYKGQME AWDGPALLLF SDGKTVGACL 480
481 DRNGLRPARY WRTSDNFVYV ASEVGVVPVD EAKVTMKGRL GPGMMIAVDL VNGQVYENTE 540
541 VKKRISSFNP YGKWIKENSR FLKPVNFKSS TVMENEEILR SQQAFGYSSE DVQMVIESMA 600
601 SQGKEPTFCM GDDIPLAGLS QRPHMLYDYF KQRFAQVTNP AIDPLREGLV MSLEVNIGKR 660
661 GNILELGPEN ASQVILSNPV LNEGALEELM KDQYLKPKVL STYFDIRKGV EGSLQKALYY 720
721 LCEAADDAVR SGSQLLVLSD RSDRLEPTRP SIPIMLAVGA VHQHLIQNGL RMSASIVADT 780
781 AQCFSTHHFA CLVGYGASAV CPYLALETCR QWRLSNKTVA FMRNGKIPTV TIEQAQKNYT 840
841 KAVNAGLLKI LSKMGISLLS SYCGAQIFEI YGLGQDVVDL AFTGSVSKIS GLTFDELARE 900
901 TLSFWVKAFS EDTTKRLENF GFIQFRPGGE YHSNNPEMSK LLHKAVREKS ETAYAVYQQH 960
961 LSNRPVNVLR DLLEFKSDRA PIPVGKVEPA VAIVQRFCTG GMSLGAISRE THEAIAIAMN1020
1021 RIGGKSNSGE GGEDPIRWKP LTDVVDGYSP TLPHLKGLQN GDIATSAIKQ VASGRFGVTP1080
1081 TFLVNADQLE IKVAQGAKPG EGGQLPGKKV SAYIARLRSS KPGVPLISPP PHHDIYSIED1140
1141 LAQLIFDLHQ INPNAKVSVK LVAEAGIGTV ASGVAKGNAD IIQISGHDGG TGASPISSIK1200
1201 HAGGPWELGL TETHQTLIAN GLRERVILRV DGGLKSGVDV LMAAAMGADE YGFGSLAMIA1260
1261 TGCVMARICH TNNCPVGVAS QREELRARFP GVPGDLVNYF LYVAEEVRGI LAQLGYNSLD1320
1321 DIIGRTELLR PRDISLVKTQ HLDLSYLLSS VGTPSLSSTE IRKQEVHTNG PVLDDDILAD1380
1381 PLVIDAIENE KVVEKTVKIC NVDRAACGRV AGVIAKKYGD TGFAGQVNLT FLGSAGQSFG1440
1441 CFLIPGMNIR LIGESNDYVG KGMAGGEIVV TPVEKIGFVP EEATIVGNTC LYGATGGQIF1500
1501 ARGKAGERFA VRNSLAEAVV EGTGDHCCEY MTGGCVVVLG KVGRNVAAGM TGGLAYLLDE1560
1561 DDTLLPKINR EIVKIQRVTA PAGELQLKSL IEAHVEKTGS SKGATILNEW EKYLPLFWQL1620
1621 VPPSEEDTPE ASAAYVRTST GEVTFQSA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
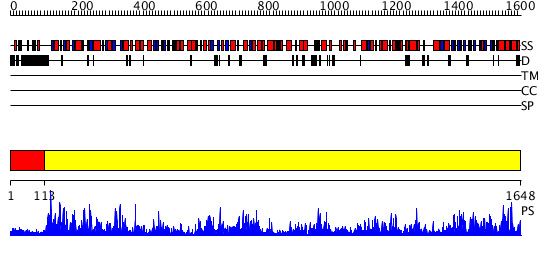
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..112] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [113..1648] | 1000.0 | Alpha subunit of glutamate synthase, C-terminal domain; Alpha subunit of glutamate synthase, central and FMN domains; Alpha subunit of glutamate synthase, N-terminal domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.61 |
Source: Reynolds et al. (2008)