













| General Information: |
|
| Name(s) found: |
DRL37_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 843 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA]
|
| Biological Process: |
apoptosis
[IEA]
defense response [ISS] |
| Molecular Function: |
ATP binding
[IEA]
protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNCCWQVVEP CYKSALSYLC VKVGNICMLK ENLVLLKSAF DELKAEKEDV VNRVNAGELK 60
61 GGQRLAIVAT WLSQVEIIEE NTKQLMDVAS ARDASSQNAS AVRRRLSTSG CWFSTCNLGE 120
121 KVFKKLTEVK SLSGKDFQEV TEQPPPPVVE VRLCQQTVGL DTTLEKTWES LRKDENRMLG 180
181 IFGMGGVGKT TLLTLINNKF VEVSDDYDVV IWVESSKDAD VGKIQDAIGE RLHICDNNWS 240
241 TYSRGKKASE ISRVLRDMKP RFVLLLDDLW EDVSLTAIGI PVLGKKYKVV FTTRSKDVCS 300
301 VMRANEDIEV QCLSENDAWD LFDMKVHCDG LNEISDIAKK IVAKCCGLPL ALEVIRKTMA 360
361 SKSTVIQWRR ALDTLESYRS EMKGTEKGIF QVLKLSYDYL KTKNAKCFLY CALFPKAYYI 420
421 KQDELVEYWI GEGFIDEKDG RERAKDRGYE IIDNLVGAGL LLESNKKVYM HDMIRDMALW 480
481 IVSEFRDGER YVVKTDAGLS QLPDVTDWTT VTKMSLFNNE IKNIPDDPEF PDQTNLVTLF 540
541 LQNNRLVDIV GKFFLVMSTL VVLDLSWNFQ ITELPKGISA LVSLRLLNLS GTSIKHLPEG 600
601 LGVLSKLIHL NLESTSNLRS VGLISELQKL QVLRFYGSAA ALDCCLLKIL EQLKGLQLLT 660
661 VTVNNDSVLE EFLGSTRLAG MTQGIYLEGL KVSFAAIGTL SSLHKLEMVN CDITESGTEW 720
721 EGKRRDQYSP STSSSEITPS NPWFKDLSAV VINSCIHLKD LTWLMYAANL ESLSVESSPK 780
781 MTELINKEKA QGVGVDPFQE LQVLRLHYLK ELGSIYGSQV SFPKLKLNKV DIENCPNLHQ 840
841 RPL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
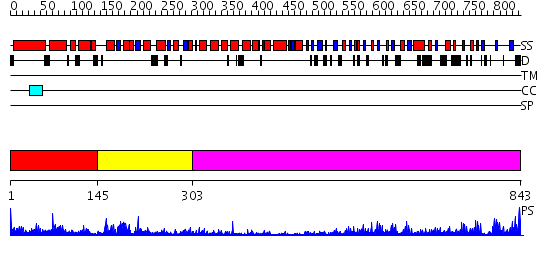
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..144] | 2.683989 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [145..302] | 6.0 | Structure of the apoptotic protease-activating factor 1 bound to ADP | |
| 3 | View Details | [303..843] | 22.0 | The molecular structure of toll-like receptor 3 ligand binding domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)