













| General Information: |
|
| Name(s) found: |
DRL31_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 874 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: |
apoptosis
[IEA]
defense response [ISS] |
| Molecular Function: |
ATP binding
[IEA]
protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGACFSVAIS CDQAVNNLTS CLSRNQNRFR NLVDHVAALK KTVRQLEARR DDLLKRIKVQ 60
61 EDRGLNLLDE VQQWLSEVES RVCEAHDILS QSDEEIDNLC CGQYCSKRCK YSYDYSKSVI 120
121 NKLQDVENLL SKGVFDEVAQ KGPIPKVEER LFHQEIVGQE AIVESTWNSM MEVGVGLLGI 180
181 YGMGGVGKTT LLSQINNKFR TVSNDFDIAI WVVVSKNPTV KRIQEDIGKR LDLYNEGWEQ 240
241 KTENEIASTI KRSLENKKYM LLLDDMWTKV DLANIGIPVP KRNGSKIAFT SRSNEVCGKM 300
301 GVDKEIEVTC LMWDDAWDLF TRNMKETLES HPKIPEVAKS IARKCNGLPL ALNVIGETMA 360
361 RKKSIEEWHD AVGVFSGIEA DILSILKFSY DDLKCEKTKS CFLFSALFPE DYEIGKDDLI 420
421 EYWVGQGIIL GSKGINYKGY TIIGTLTRAY LLKESETKEK VKMHDVVREM ALWISSGCGD 480
481 QKQKNVLVVE ANAQLRDIPK IEDQKAVRRM SLIYNQIEEA CESLHCPKLE TLLLRDNRLR 540
541 KISREFLSHV PILMVLDLSL NPNLIELPSF SPLYSLRFLN LSCTGITSLP DGLYALRNLL 600
601 YLNLEHTYML KRIYEIHDLP NLEVLKLYAS GIDITDKLVR QIQAMKHLYL LTITLRNSSG 660
661 LEIFLGDTRF SSYTEGLTLD EQSYYQSLKV PLATISSSRF LEIQDSHIPK IEIEGSSSNE 720
721 SEIVGPRVRR DISFINLRKV RLDNCTGLKD LTWLVFAPHL ATLYVVCLPD IEHIISRSEE 780
781 SRLQKTCELA GVIPFRELEF LTLRNLGQLK SIYRDPLLFG KLKEINIKSC PKLTKLPLDS 840
841 RSAWKQNVVI NAEEEWLQGL QWEDVATKER FFPS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
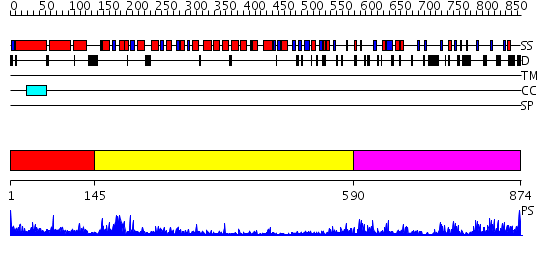
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..144] | 4.30103 | Crystal Structure Analysis of ClpB | |
| 2 | View Details | [145..589] | 13.30103 | Structure of the apoptotic protease-activating factor 1 bound to ADP | |
| 3 | View Details | [590..874] | 21.39794 | The molecular structure of toll-like receptor 3 ligand binding domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)