













| General Information: |
|
| Name(s) found: |
CP74A_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 518 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA][ISS]
mitochondrion [IDA] membrane [IDA] chloroplast envelope [IDA] plastoglobule [IDA] chloroplast thylakoid membrane [IDA] thylakoid [IDA] |
| Biological Process: |
oxylipin metabolic process
[IDA]
epoxygenase P450 pathway [ISS] jasmonic acid biosynthetic process [IDA][TAS] response to wounding [IEP][IMP] response to jasmonic acid stimulus [IMP] response to fungus [IEP] defense response [TAS] |
| Molecular Function: |
oxygen binding
[ISS]
hydro-lyase activity [IDA] allene oxide synthase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASISTPFPI SLHPKTVRSK PLKFRVLTRP IKASGSETPD LTVATRTGSK DLPIRNIPGN 60
61 YGLPIVGPIK DRWDYFYDQG AEEFFKSRIR KYNSTVYRVN MPPGAFIAEN PQVVALLDGK 120
121 SFPVLFDVDK VEKKDLFTGT YMPSTELTGG YRILSYLDPS EPKHEKLKNL LFFLLKSSRN 180
181 RIFPEFQATY SELFDSLEKE LSLKGKADFG GSSDGTAFNF LARAFYGTNP ADTKLKADAP 240
241 GLITKWVLFN LHPLLSIGLP RVIEEPLIHT FSLPPALVKS DYQRLYEFFL ESAGEILVEA 300
301 DKLGISREEA THNLLFATCF NTWGGMKILF PNMVKRIGRA GHQVHNRLAE EIRSVIKSNG 360
361 GELTMGAIEK MELTKSVVYE CLRFEPPVTA QYGRAKKDLV IESHDAAFKV KAGEMLYGYQ 420
421 PLATRDPKIF DRADEFVPER FVGEEGEKLL RHVLWSNGPE TETPTVGNKQ CAGKDFVVLV 480
481 ARLFVIEIFR RYDSFDIEVG TSPLGSSVNF SSLRKASF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
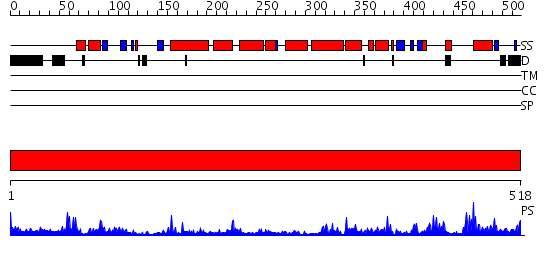
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..518] | 85.045757 | Crystal Structure of Human Drug Metabolizing Cytochrome P450 2C8 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)