













| General Information: |
|
| Name(s) found: |
COP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 675 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][IMP]
nuclear ubiquitin ligase complex [TAS] CUL4 RING ubiquitin ligase complex [ISS] |
| Biological Process: |
photoperiodism, flowering
[IMP]
DNA repair [IMP] anthocyanin metabolic process [IMP] photomorphogenesis [IGI][TAS] regulation of stomatal movement [IGI][IMP] shade avoidance [IMP] skotomorphogenesis [TAS] |
| Molecular Function: |
ubiquitin-protein ligase activity
[IDA]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEEISTDPVV PAVKPDPRTS SVGEGANRHE NDDGGSGGSE IGAPDLDKDL LCPICMQIIK 60
61 DAFLTACGHS FCYMCIITHL RNKSDCPCCS QHLTNNQLYP NFLLDKLLKK TSARHVSKTA 120
121 SPLDQFREAL QRGCDVSIKE VDNLLTLLAE RKRKMEQEEA ERNMQILLDF LHCLRKQKVD 180
181 ELNEVQTDLQ YIKEDINAVE RHRIDLYRAR DRYSVKLRML GDDPSTRNAW PHEKNQIGFN 240
241 SNSLSIRGGN FVGNYQNKKV EGKAQGSSHG LPKKDALSGS DSQSLNQSTV SMARKKRIHA 300
301 QFNDLQECYL QKRRQLADQP NSKQENDKSV VRREGYSNGL ADFQSVLTTF TRYSRLRVIA 360
361 EIRHGDIFHS ANIVSSIEFD RDDELFATAG VSRCIKVFDF SSVVNEPADM QCPIVEMSTR 420
421 SKLSCLSWNK HEKNHIASSD YEGIVTVWDV TTRQSLMEYE EHEKRAWSVD FSRTEPSMLV 480
481 SGSDDCKVKV WCTRQEASVI NIDMKANICC VKYNPGSSNY IAVGSADHHI HYYDLRNISQ 540
541 PLHVFSGHKK AVSYVKFLSN NELASASTDS TLRLWDVKDN LPVRTFRGHT NEKNFVGLTV 600
601 NSEYLACGSE TNEVYVYHKE ITRPVTSHRF GSPDMDDAEE EAGSYFISAV CWKSDSPTML 660
661 TANSQGTIKV LVLAA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
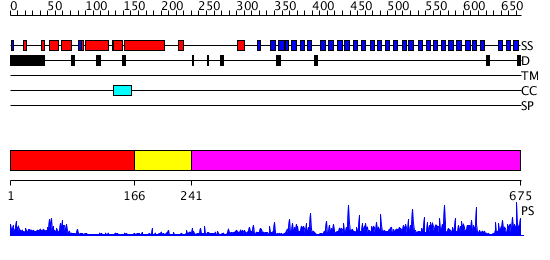
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..165] | 3.522879 | No description for 2cklB was found. | |
| 2 | View Details | [166..240] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [241..675] | 64.69897 | F-box/WD-repeat protein 1 (beta-TRCP1) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)