













| General Information: |
|
| Name(s) found: |
CNX1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 670 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
Mo-molybdopterin cofactor biosynthetic process
[IDA][IMP]
auxin mediated signaling pathway [IMP] |
| Molecular Function: |
molybdenum ion binding
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEGQGCCGGG GGKTEMIPTE EALRIVFGVS KRLPPVIVSL YEALGKVLAE DIRAPDPLPP 60
61 YPASVKDGYA VVASDGPGEY PVITESRAGN DGLGVTVTPG TVAYVTTGGP IPDGADAVVQ 120
121 VEDTKVIGDV STESKRVKIL IQTKKGTDIR RVGCDIEKDA TVLTTGERIG ASEIGLLATA 180
181 GVTMVKVYPM PIVAILSTGD ELVEPTAGTL GRGQIRDSNR AMLVAAVMQQ QCKVVDLGIV 240
241 RDDRKELEKV LDEAVSSGVD IILTSGGVSM GDRDFVKPLL EEKGKVYFSK VLMKPGKPLT 300
301 FAEIRAKPTE SMLGKTVLAF GLPGNPVSCL VCFNIFVVPT IRQLAGWTSP HPLRVRLRLQ 360
361 EPIKSDPIRP EFHRAIIKWK DNDGSGTPGF VAESTGHQMS SRLLSMRSAN ALLELPATGN 420
421 VLSAGSSVSA IIVSDISAFS IDKKASLSEP GSIRKEKKYD EVPGPEYKVA ILTVSDTVSA 480
481 GAGPDRSGPR AVSVVDSSSE KLGGAKVVAT AVVPDEVERI KDILQKWSDV DEMDLILTLG 540
541 GTGFTPRDVT PEATKKVIER ETPGLLFVMM QESLKITPFA MLSRSAAGIR GSTLIINMPG 600
601 NPNAVAECME ALLPALKHAL KQIKGDKREK HPKHIPHAEA TLPTDTWDQS YKSAYETGEK 660
661 KEEAGCSCTH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
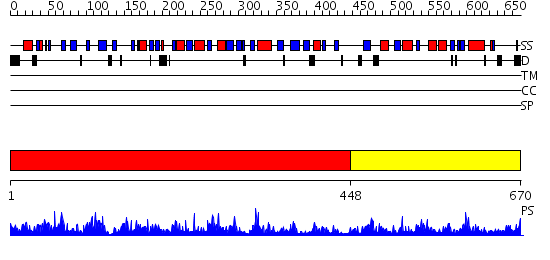
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..447] | 109.0 | Structural basis of dynamic glycine receptor clustering | |
| 2 | View Details | [448..670] | 34.69897 | Plant CNX1 G domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)