













| General Information: |
|
| Name(s) found: |
CNG13_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 696 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA]
|
| Biological Process: |
ion transport
[IEA]
|
| Molecular Function: |
ion channel activity
[ISS]
calmodulin binding [ISS] cyclic nucleotide binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAFGRNNRVR FRDWISEGTE YGYGRNKARP SLNTVLKNVR RGLKKPLSFG SHNKKRDSNS 60
61 STTTQKNIIN PQGSFLQNWN KIFLFASVIA LAIDPLFFYI PIVDGERHCL NLHRNLEIAA 120
121 SVLRTFIDAF YIIHIVFQFR TAYISPSSRV FGRGELVDDP KAIAIKYLSS YFIIDLLSIL 180
181 PLPQLVVLAV IPNVNKPVSL ITKDYLITVI FTQYIPRILR IYPLYTEVTR TSGIVTETAW 240
241 AGAAWNLSLY MLASHVFGAL WYLISVERED RCWREACEKI PEVCNFRFLY CDGNSSVRND 300
301 FLTTSCPFIN PDDITNSTVF NFGIFTDALK SGIVESDDFW KKFFYCFWWG LRNLSALGQN 360
361 LNTSKFVGEI IFAVSICISG LVLFALLIGN MQKYLESTTV REEEMRVRKR DAEQWMSHRM 420
421 LPDDLRKRIR RYEQYKWQET RGVEEENLLR NLPKDLRRDI KRHFCLDLLK KVPLFEIMDE 480
481 QLLDAVCDKL KPVLYTENSY AIREGDPVEE MLFVMRGKLM SATTNGGRTG FFNAVYLKPS 540
541 DFCGEDLLTW ALDPQSSSHF PISTRTVQAL TEVEAFALAA DDLKLVASQF RRLHSKQLQH 600
601 TFRFYSVQWR TWGASFIQAA WRRHCRRKLA RSLTEEEDRF RNAITKRERN AASSSSLVAT 660
661 LYASRFASNA LRNLRTNNLP LLPPKPSEPD FSLRNP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
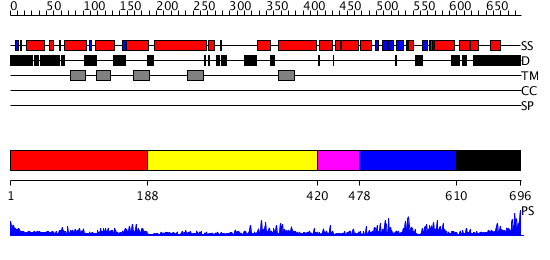
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..187] | 1.58 | Crystal structure of KvAP-33H1 Fv complex | |
| 2 | View Details | [188..419] | 10.0 | Mammalian Shaker Kv1.2 potassium channel- beta subunit complex | |
| 3 | View Details | [420..477] | 2.3 | No description for 2ptmA was found. | |
| 4 | View Details | [478..609] | 20.045757 | Solution structure of the cNMP-binding domain from Arabidopsis thaliana cyclic nucleotide-regulated ion channel | |
| 5 | View Details | [610..696] | 2.899994 | View MSA. Confident ab initio structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|