













| General Information: |
|
| Name(s) found: |
ARFE_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 902 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
membrane [IDA] |
| Biological Process: |
embryonic development
[IGI]
root development [IGI] flower development [IGI] xylem and phloem pattern formation [IMP] leaf vascular tissue pattern formation [IGI] meristem development [IGI] response to auxin stimulus [IEP] longitudinal axis specification [IMP] |
| Molecular Function: |
transcription factor activity
[ISS][TAS]
promoter binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMASLSCVED KMKTSCLVNG GGTITTTTSQ STLLEEMKLL KDQSGTRKPV INSELWHACA 60
61 GPLVCLPQVG SLVYYFSQGH SEQVAVSTRR SATTQVPNYP NLPSQLMCQV HNVTLHADKD 120
121 SDEIYAQMSL QPVHSERDVF PVPDFGMLRG SKHPTEFFCK TLTASDTSTH GGFSVPRRAA 180
181 EKLFPPLDYS AQPPTQELVV RDLHENTWTF RHIYRGQPKR HLLTTGWSLF VGSKRLRAGD 240
241 SVLFIRDEKS QLMVGVRRAN RQQTALPSSV LSADSMHIGV LAAAAHATAN RTPFLIFYNP 300
301 RACPAEFVIP LAKYRKAICG SQLSVGMRFG MMFETEDSGK RRYMGTIVGI SDLDPLRWPG 360
361 SKWRNLQVEW DEPGCNDKPT RVSPWDIETP ESLFIFPSLT SGLKRQLHPS YFAGETEWGS 420
421 LIKRPLIRVP DSANGIMPYA SFPSMASEQL MKMMMRPHNN QNVPSFMSEM QQNIVMGNGG 480
481 LLGDMKMQQP LMMNQKSEMV QPQNKLTVNP SASNTSGQEQ NLSQSMSAPA KPENSTLSGC 540
541 SSGRVQHGLE QSMEQASQVT TSTVCNEEKV NQLLQKPGAS SPVQADQCLD ITHQIYQPQS 600
601 DPINGFSFLE TDELTSQVSS FQSLAGSYKQ PFILSSQDSS AVVLPDSTNS PLFHDVWDTQ 660
661 LNGLKFDQFS PLMQQDLYAS QNICMSNSTT SNILDPPLSN TVLDDFCAIK DTDFQNHPSG 720
721 CLVGNNNTSF AQDVQSQITS ASFADSQAFS RQDFPDNSGG TGTSSSNVDF DDCSLRQNSK 780
781 GSSWQKIATP RVRTYTKVQK TGSVGRSIDV TSFKDYEELK SAIECMFGLE GLLTHPQSSG 840
841 WKLVYVDYES DVLLVGDDPW EEFVGCVRCI RILSPTEVQQ MSEEGMKLLN SAGINDLKTS 900
901 VS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
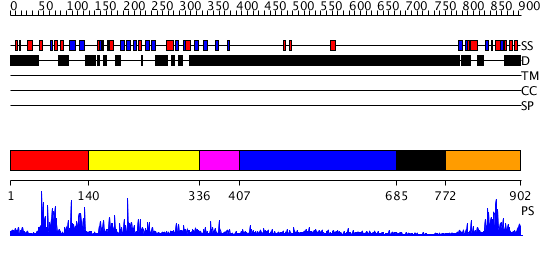
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..139] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [140..335] | 33.221849 | Solution Structure of the B3 DNA-Binding Domain of RAV1 | |
| 3 | View Details | [336..406] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [407..684] | 2.310996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [685..771] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [772..902] | 5.197976 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)