













| General Information: |
|
| Name(s) found: |
ARFB_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 859 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
chloroplast [IEA] |
| Biological Process: |
negative regulation of transcription, DNA-dependent
[TAS]
leaf senescence [IMP] negative regulation of cell proliferation [IMP] floral organ abscission [IMP] ovule development [IMP] positive regulation of flower development [IMP] fruit dehiscence [IMP] |
| Molecular Function: |
transcription factor activity
[ISS]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASSEVSMKG NRGGDNFSSS GFSDPKETRN VSVAGEGQKS NSTRSAAAER ALDPEAALYR 60
61 ELWHACAGPL VTVPRQDDRV FYFPQGHIEQ VEASTNQAAE QQMPLYDLPS KLLCRVINVD 120
121 LKAEADTDEV YAQITLLPEA NQDENAIEKE APLPPPPRFQ VHSFCKTLTA SDTSTHGGFS 180
181 VLRRHADECL PPLDMSRQPP TQELVAKDLH ANEWRFRHIF RGQPRRHLLQ SGWSVFVSSK 240
241 RLVAGDAFIF LRGENGELRV GVRRAMRQQG NVPSSVISSH SMHLGVLATA WHAISTGTMF 300
301 TVYYKPRTSP SEFIVPFDQY MESVKNNYSI GMRFKMRFEG EEAPEQRFTG TIVGIEESDP 360
361 TRWPKSKWRS LKVRWDETSS IPRPDRVSPW KVEPALAPPA LSPVPMPRPK RPRSNIAPSS 420
421 PDSSMLTREG TTKANMDPLP ASGLSRVLQG QEYSTLRTKH TESVECDAPE NSVVWQSSAD 480
481 DDKVDVVSGS RRYGSENWMS SARHEPTYTD LLSGFGTNID PSHGQRIPFY DHSSSPSMPA 540
541 KRILSDSEGK FDYLANQWQM IHSGLSLKLH ESPKVPAATD ASLQGRCNVK YSEYPVLNGL 600
601 STENAGGNWP IRPRALNYYE EVVNAQAQAQ AREQVTKQPF TIQEETAKSR EGNCRLFGIP 660
661 LTNNMNGTDS TMSQRNNLND AAGLTQIASP KVQDLSDQSK GSKSTNDHRE QGRPFQTNNP 720
721 HPKDAQTKTN SSRSCTKVHK QGIALGRSVD LSKFQNYEEL VAELDRLFEF NGELMAPKKD 780
781 WLIVYTDEEN DMMLVGDDPW QEFCCMVRKI FIYTKEEVRK MNPGTLSCRS EEEAVVGEGS 840
841 DAKDAKSASN PSLSSAGNS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
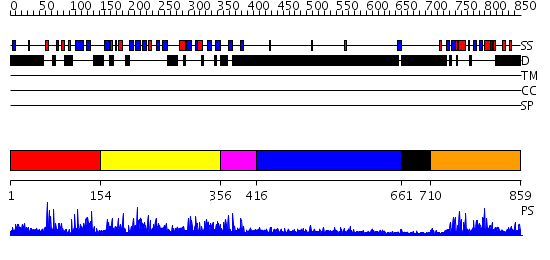
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..153] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [154..355] | 38.69897 | Solution Structure of the B3 DNA-Binding Domain of RAV1 | |
| 3 | View Details | [356..415] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [416..660] | 7.303997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [661..709] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [710..859] | 1.06 | Bud emergence mediator Bemp1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)