













| General Information: |
|
| Name(s) found: |
AAT3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 449 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[IDA]
membrane [IDA] plastid [IDA] |
| Biological Process: |
leaf senescence
[IEP]
|
| Molecular Function: |
L-aspartate:2-oxoglutarate aminotransferase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKTTHFSSSS SSDRRIGALL RHLNSGSDSD NLSSLYASPT SGGTGGSVFS HLVQAPEDPI 60
61 LGVTVAYNKD PSPVKLNLGV GAYRTEEGKP LVLNVVRKAE QQLINDRTRI KEYLPIVGLV 120
121 EFNKLSAKLI LGADSPAIRE NRITTVECLS GTGSLRVGGE FLAKHYHQKT IYITQPTWGN 180
181 HPKIFTLAGL TVKTYRYYDP ATRGLNFQGL LEDLGAAAPG SIVLLHACAH NPTGVDPTIQ 240
241 QWEQIRKLMR SKGLMPFFDS AYQGFASGSL DTDAKPIRMF VADGGECLVA QSYAKNMGLY 300
301 GERVGALSIV CKSADVAGRV ESQLKLVIRP MYSSPPIHGA SIVAVILRDK NLFNEWTLEL 360
361 KAMADRIISM RKQLFEALRT RGTPGDWSHI IKQIGMFTFT GLNPAQVSFM TKEYHIYMTS 420
421 DGRISMAGLS SKTVPHLADA IHAVVTKAV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
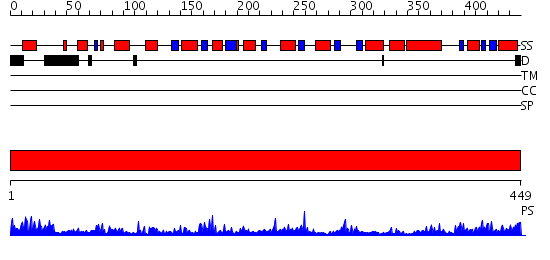
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..449] | 87.154902 | DOMAIN CLOSURE IN MITOCHONDRIAL ASPARTATE AMINOTRANSFERASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)