













| General Information: |
|
| Name(s) found: |
YKCC_BACSU
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Bacillus subtilis |
| Length: | 323 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRHIQYSIV VPVYNEELVI HETYQRLKEV MDQTKENYEL LFVNDGSKDR SIEILREHSL 60
61 IDPRVKIIDF SRNFGHQIAI TAGMDYAQGN AIVVIDADLQ DPPELILEMI EKWKEGYEVV 120
121 YAVRTKRKGE TFFKKQTAAM FYRLLSGMTD IDIPIDTGDF RLMDRKVCDE MKRLKEKNPF 180
181 VRGLVSWVGS KQTAVEYVRD ERLAGETKYP LKKMLKLSMD GITTFSHKPL KLASYAGILM 240
241 SGTGFLYMFI VLYLKLFTDS TITGWSSLIV IQLLFSGIVL LILGVIGEYI GRIYDEAKDR 300
301 PLYIVQKSYG IENKRLYRDQ HMS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
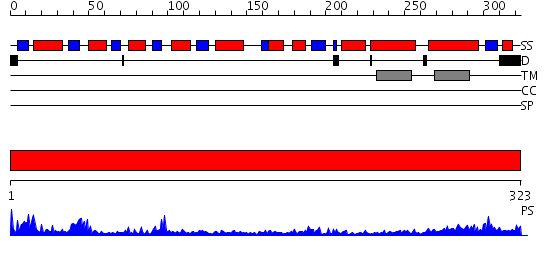
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..323] | 20.0 | The Crystal Structure of UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase-T1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|