













| General Information: |
|
| Name(s) found: |
CE35517
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 671 amino acids |
Gene Ontology: |
|
| Cellular Component: |
phosphoribosylaminoimidazole carboxylase complex
[IEA cytoplasm [IEA |
| Biological Process: |
sodium ion transport
[IEA gluconeogenesis [IEA metabolic process [IEA peptidoglycan biosynthetic process [IEA protein modification process [IEA 'de novo' IMP biosynthetic process [IEA purine base biosynthetic process [IEA |
| Molecular Function: |
oxaloacetate decarboxylase activity
[IEA ATP binding [IEA D-alanine-D-alanine ligase activity [IEA phosphoribosylaminoimidazole carboxylase activity [IEA pyruvate carboxylase activity [IEA biotin binding [IEA ligase activity [IEA hydroxymethyl-, formyl- and related transferase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLGVFQKRCA STAPLIRPIQ RVLVANRGEI AIRVQNTARK MGIETVAVFS DADRNSLFVK 60
61 KADKAYHIGP PLAAESYLNM DKIINSALRS GAQAIHPGYG FLSENAGFAE KCAQAGLVFI 120
121 GPPAQAIRDM GAKNVSKQIM EDAKVPVVKG FHGEDQSDAN LKKKSAEIGY PVMLKAVYGG 180
181 GGKGMRIAWN EAEFEEKLAS ARNEAKKSFG NDEMLVEKFV EKPRHVEVQV FGDHHGNYVH 240
241 LWERDCSVQR RHQKIIEEAP APNMEHDTRV KLGESAVRAA AAVGYVGAGT VEFIMDPRGE 300
301 FYFMEMNTRL QVEHPVSEAI TGTDLVEWQL RVAQGEKLPL KQSEIPLNGH AFECRVYAED 360
361 TRKGAFMPTA GRLNYVDFPE DARIDTGVVS GDEVSIHYDP MIAKVVVWGQ DRAVAAAKLE 420
421 SALARTRISG LPTNIDFVRR VLAHPEFAAG NVYTDFIPDH QKELFAESET PAEIYVESAV 480
481 AHALAALQKS DAGVFQNLSF FRMNLSPKYI FKIDGKDVSI RFDSDSRLTV SYDGNSYETL 540
541 LNDIESTTDG SFKFTLEANG RRWSTVVKNL GNSLMVNGVG QNEYETPQIH ETFDSLSGGA 600
601 ASHSAVAPMP GIIEKILVKP GEQVTTGQAL VVMTAMKMEY IIRAPEDSTV EHIKCQAGKN 660
661 VPKNAVLVQF A |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
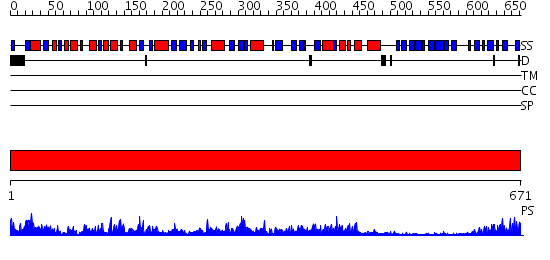
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..671] | 147.0 | No description for 2qf7A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)