













| General Information: |
|
| Name(s) found: |
CE35176
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 470 amino acids |
Gene Ontology: |
|
| Cellular Component: |
HslUV protease complex
[IEA cytoplasm [IEA |
| Biological Process: |
protein transport
[IEA |
| Molecular Function: |
nucleotide binding
[IEA ATPase activity [IEA ATP binding [IEA protein binding [IEA nucleoside-triphosphatase activity [IEA HslUV protease activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFSKQFARKT PPYPTQIAEY LDKFVVGQKK AKKTLAVGVY QHYRRLEHNI ETGASSIYQT 60
61 HAQTSSSGKS LSNDGMDPKM PRGVFYQDEL RLGQMASSEL RNSIMQQQSN NQPPSPAQSP 120
121 RNAAPTFRAL PEKEQSVRLE KSNVLLVGPS GVGKTFLTQT LARVLDVPIA LCDCTSMTQA 180
181 GYVGEDVESV IQKLVQAAGG NVEKAQQGIV FLDEVDKIAA AHEGHSAAYR DVSGEGVQHA 240
241 LLKLVEGTVV NVKSGKKGMG SQQDQVQIDT TDILFIASGA FSNLDKIVGR RLDKKALGFG 300
301 TSSGNVRISG DDSNSEVMRK RDELLSKADQ GDLISFGMVP ELVGRFPVLV PFHSFDKQML 360
361 VRVMTEPQNS LLAQLKLQFG IDNVDLSFSA EALEQVAQLA LDRKTGARAL RSILEAALLE 420
421 AKFTVPGSDI ESVHVSREAI LGEKEVEYSR RKSQVVEEED VSPMKKSAVA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
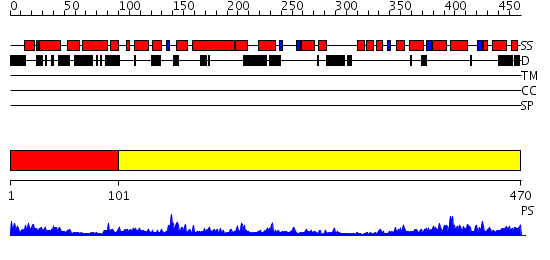
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..100] | 3.64 | Crystal structure of helicobacter pylori ClpX | |
| 2 | View Details | [101..470] | 64.69897 | Crystal structure of helicobacter pylori ClpX |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)