













| General Information: |
|
| Name(s) found: |
fbxa-1 /
CE33881
[WormBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 766 amino acids |
Gene Ontology: |
|
| Cellular Component: |
protein serine/threonine phosphatase complex
[IEA |
| Biological Process: |
protein amino acid dephosphorylation
[IEA |
| Molecular Function: |
protein serine/threonine phosphatase activity
[IEA catalytic activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVQTSEPMAR TPIQFGENMR ITVAASQGGR RYMEDRCVIH TERINNGLLD WTFVGVFDGH 60
61 GGEHASEYVR RHLLMNITKN QKFESNSDED ILEAIRQGFL MTHEQMRHVY DEWPYTASGY 120
121 PSTAGTTVSC VFIRNGKLYT GHVGDSAIFL GTVENGELHS RPLTTDHKPE SVHEQLRIAK 180
181 AGGETAVKSG VTRVVWKRPQ KMSQFMMMTS NSNEQKHHQN PQIMENIPFL SVARSLGDLW 240
241 SYNEKTNMFI VSPEPDLGVH RLTGNDFCLV LASDGMTNVM TGDQAISIVF KEEEMVEIHE 300
301 EINRNHSRCV LRSALQKWRS LRADNVTIAT VIFDIEPIKY TDAEMLMKVG KYVNASQVLT 360
361 DRPDAMLKVS KNENILLSTQ RTPIIYNGSK DVNFCRVTYK GPGFRTHEDE LLEERRNLMS 420
421 KFGGVDGGAA IAIAGGPPVD ARRGDTTGKD ENNVKYDVNG EEDDIINDIT TEEEASQEID 480
481 LSLRLVQRST AVTIIESPNR PPRPHAPRPV FIDDSIIEEE NDGNLDEKSL IGVQITPSRT 540
541 SPGHCTVRRV KAEAVRKLRT PKMSTMDESS SSSTPPDRRI TRSASKGLKT PPTPEPPRGI 600
601 IGVNPNLTAD AVLLLTPRRR RSTRIQNSCA SAAAPGAASL STEKLTQSSR KRARTCAVGA 660
661 ASGVTPVVAS LTLQDHQLLR PHTHAPPPTP SRSAPSSPSS KKCEISSSLK TSKSAKASSL 720
721 SKLDDGIIRV SEIADEEDDD DVTMMRGPPS KMRRFFGYMK KMVWGK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
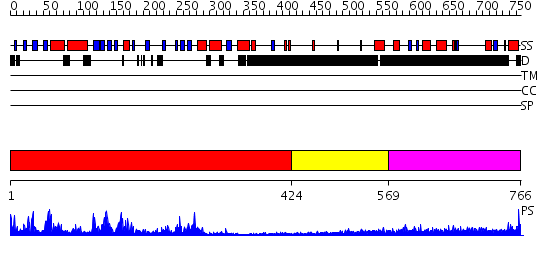
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..423] | 88.221849 | CRYSTAL STRUCTURE OF THE PROTEIN SERINE/THREONINE PHOSPHATASE 2C AT 2 A RESOLUTION | |
| 2 | View Details | [424..568] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [569..766] | 6.200994 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)