













| General Information: |
|
| Name(s) found: |
CE33176
[WormBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 444 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
arginine metabolic process
[IEA ectoine biosynthetic process [IEA tetrapyrrole biosynthetic process [IEA biosynthetic process [IEA biotin biosynthetic process [IEA gamma-aminobutyric acid metabolic process [IEA |
| Molecular Function: |
pyridoxal phosphate binding
[IEA 4-aminobutyrate transaminase activity [IEA transaminase activity [IEA glutamate-1-semialdehyde 2,1-aminomutase activity [IEA diaminobutyrate-pyruvate transaminase activity [IEA adenosylmethionine-8-amino-7-oxononanoate transaminase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQRVQALRPL LPKGHVTYYK DQLLITKGEK QFLFDSNGKK YLDFFGGIVT VSVGHCHPKI 60
61 NAALTEQAQK LWHTTSIYHT EPIYEYAEKL LSKFPSKLNS VFFVNSGSEA NDLALALARN 120
121 YTGRFDVISM RNGYHGMTQT VLGATNLGNW KPVFPHGFNI FKSLNADPYR GIFGGSNCRD 180
181 SPIQVKNRKC DCKPGSCQAS DKYIEQFDDM LLHDFSHSSG PAAFLIESIQ GVGGTVQYPH 240
241 GYLKKSYESV QKRGGLAIAD EVQTGFGRLG SHFWGFESQD ALPDMVTMAK GIGNGFPLGA 300
301 VVTSKEIADS FNKSLYFNTY GGNPLASVVG KAVLEVIEEE KLQENSAVVG DYFLKQLAAI 360
361 DDATIGDVRG KGLMIGVELI DEQGKPLAAA KTAAIFEDTK NQGLLIGKGG IHGNVLRIKP 420
421 PMCITKKDVD FAVDIIAKSI KQYK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
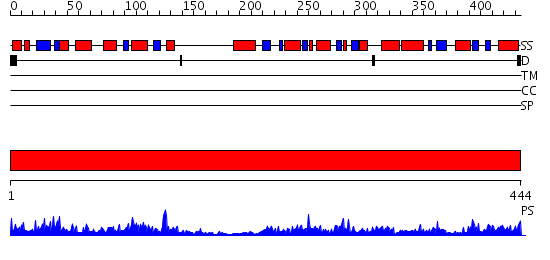
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..444] | 100.0 | The structure of gamma-aminobutyrate aminotransferase mutant: E211S |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)