













| General Information: |
|
| Name(s) found: |
nhr-88 /
CE33143
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 593 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
DNA integration
[IEA regulation of transcription, DNA-dependent [IEA transposition, DNA-mediated [IEA |
| Molecular Function: |
DNA binding
[IEA transcription factor activity [IEA zinc ion binding [IEA transposase activity [IEA steroid hormone receptor activity [IEA sequence-specific DNA binding [IEA steroid binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFAPSTVYTY GHHGSGTDED DGQQVMPGMD NGDGQCMVCG DRSAGKHYGV MACYGCKGFF 60
61 RRTIRSQQTY TCRFTQKCAI DKDQRNACRY CRFQRCLTVG MEPEAIRPDR DVIGKQKNPR 120
121 KKKMKSDTNS GSIDASLPSP NGCDSPVSNN EDVILSFLLD VEDQATCGNN HITMPIGISM 180
181 MMKNDPDFDV STLFHSQYVR NQESFPITYA VGRTASVEQL IAALRRYVLS AVHWIDALFN 240
241 LAHLTEIHDK TTLLKSVIGP FTIFNIAART AQISDGDLIC LCNKSTINRQ PARHLLDTNL 300
301 VGNNFVGRVI DDLVFPTKKL ALTNPEITIL SALIILDPDA RGLSQETSQA LLGIRDRVQN 360
361 ALFNLIRDNS NNMTSVTSRF GNLLLLFPPL AKLSSLIGEN VQLAKMFGIP IDSLLVELYV 420
421 DADSPELISQ GSHARERSDV STQTASLEGA SASMTPDVTS SEFVKMEQNE SPSSSEEPMC 480
481 IGAPATSSAN AVATNLAADL LGLLPNFDAT GLDLASAAAA AATSVNYSHF YGFGNLGGSL 540
541 DDSSSSGTGS VGSFAPHSAP PISNNVNPFA VHGFFDNSAP NTGVVHNQPF RFV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
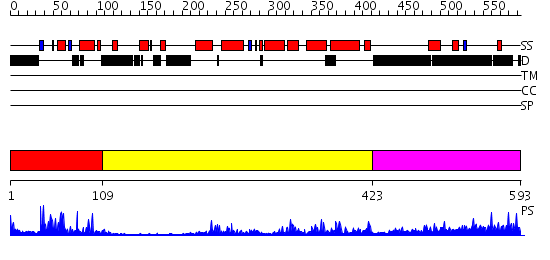
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..108] | 30.522879 | Human Liver Receptor Homologue DNA-Binding Domain (hLRH-1 DBD) in Complex with dsDNA from the hCYP7A1 Promoter | |
| 2 | View Details | [109..422] | 32.522879 | hemipteran ecdysone receptor ligand-binding domain complexed with ponasterone A | |
| 3 | View Details | [423..593] | 16.302997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)