













| General Information: |
|
| Name(s) found: |
CE31383
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 461 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
carbohydrate metabolic process
[IEA biosynthetic process [IEA |
| Molecular Function: |
ATP binding
[IEA nucleotidyltransferase activity [IEA catalytic activity [IEA phosphotransferase activity, alcohol group as acceptor [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSKYGLLVL NSKNVQKLKQ LLTSAAAALS SKLYVRLNPD EPRTDELVSK IYLTSASSCP 60
61 KLDVRLVVSE IRGNPENYVL IGEEKLLENA EKLAAAKKYK KVVLGGTFDR LHNGHKVLLN 120
121 KAAELASDVI VVGVTDKDMI IKKSLFEMIE PVEFRIKKVV DFVEDISGTA KCLAEPIIDP 180
181 FGPSTRIKDL EAIVVSRETI KGADAVNKKR NEQGMSQLDV IIVELVEGSD EILKETKISS 240
241 SSRRREDLGH LLRPARAVPR ETGAPYIIGL AGGIASGKSH IGKYLRETHN FDVIDCDKLA 300
301 HTCYERGSSL NRKIGEHFGG DVVVDGVVDR RKLGTIVFSD KVKLRELSEL VWPEVKEKAM 360
361 EIVKKSTAKV VVIEAAALIE AGWHKTLAET WTVFVPADEA VRRVVARDNL TEDQAKDRMS 420
421 SQISNKERLD NSNVALCSLW AYEETHAQVD RAVEELMKRI L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
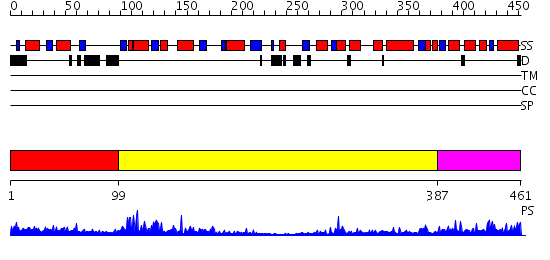
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..98] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [99..386] | 15.0 | Transcriptional regulator NadR, NMN-adenylyltransferase domain; Transcriptional regulator NadR, ribosylnicotinamide kinase domain | |
| 3 | View Details | [387..461] | 31.0 | Crystal structure of Bifunctional coenzyme A synthase (CoA synthase): (18044849) from MUS MUSCULUS at 1.70 A resolution |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.54 |
Source: Reynolds et al. (2008)