













| General Information: |
|
| Name(s) found: |
CE31247
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 651 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
carbohydrate metabolic process
[IEA D-ribose metabolic process [IEA |
| Molecular Function: |
thiamin pyrophosphate binding
[IEA nucleotidyltransferase activity [IEA ribokinase activity [IEA phosphotransferase activity, alcohol group as acceptor [IEA carbon-carbon lyase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCSRFFGQKR SFSKFLQISP QVKEALASGE GVVALESTVI THGLPYPHNL STARSLEQKV 60
61 RSSGSHPATI ALFDGKIHVG LDDEKLELLA SSQNAVKVSS RDIAKTLIKK EVGGTTVAST 120
121 MKIAHAAGIS VFATGGIGGV HRGADQTFDV SADLQELSQT PVCVVCSGVK SILDIPKTVE 180
181 YLETHSVNCI VYGQENVFPS FFTRTSDRKA QFCTESLEEV VHLLKTSKSL GLPYGTILAC 240
241 PIPEKYAADG DAIQKAIDQA VQEAIEQNIA SQSVTPFILA RVNELTQGAS MATNIALLEN 300
301 NASIAGRLAA KLCDRRPLTI SQSQPTASTT RKPKVVSIGA TIVDFEAITS EDVKDDGGSY 360
361 NGQVVQRMGG VARNHADALA RLGCDSVFIS AIGDDNNGHF FRQNSHKIDI NRVKIISNKP 420
421 TCTYLAVNVK GNVKYGIVTC EPLLSVITPS LVESNEDALE SSDFILLDSN LSVPVMARVL 480
481 EIAKKHDKQV WLEPTDIDKV KKVFDTGLVG AVTAASPNAN EFLKWAKLCH VSVDPSVIDT 540
541 ADGVLELIEK EKTKLLLNTS IFIVTLANKG SAVVYRNKLG QLEFQSLPPP LQMNKVVSVS 600
601 GAGDSFNSGV IAGLAHNKTV VESLQIGQEC ARLTLQTSLA ISDAITPNLL A |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
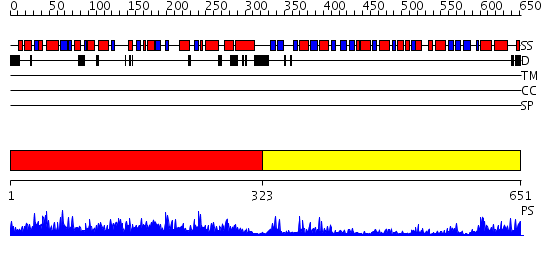
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..322] | 76.69897 | Crystal structure of conserved hypothetical protein possibly involved in carbohydrate metabolism (TM1464) from Thermotoga maritima at 1.90 A resolution | |
| 2 | View Details | [323..651] | 53.0 | Crystal structure of Ribokinase (TM0960) from Thermotoga maritima at 2.15 A resolution |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)